- Search by keyword
- Search by citation
Page 1 of 17

PSMD1 and PSMD2 regulate HepG2 cell proliferation and apoptosis via modulating cellular lipid droplet metabolism
Obesity and nonalcoholic steatohepatitis (NASH) are well-known risk factors of hepatocellular carcinoma (HCC). The lipid-rich environment enhances the proliferation and metastasis abilities of tumor cells. Pre...
- View Full Text
The effect of BACE1-AS on β-amyloid generation by regulating BACE1 mRNA expression
The BACE1 antisense transcript (BACE1-AS) is a conserved long noncoding RNA (lncRNA). The level of BACE1-AS is significantly increased and the level of the BACE1 mRNA is slightly increased in subjects with AD....
Overlapping transcriptional expression response of wheat zinc-induced facilitator-like transporters emphasize important role during Fe and Zn stress
Hexaploid wheat is an important cereal crop that has been targeted to enhance grain micronutrient content including zinc (Zn) and iron (Fe). In this direction, modulating the expression of plant transporters i...
MiR-32-5p influences high glucose-induced cardiac fibroblast proliferation and phenotypic alteration by inhibiting DUSP1
The current study aimed to investigate the effects of miR-32-5p on cardiac fibroblasts (CFs) that were induced with high levels of glucose; we also aimed to identify the potential mechanisms involved in the re...
Correction to: A protocol for custom CRISPR Cas9 donor vector construction to truncate genes in mammalian cells using pcDNA3 backbone
The original article [1] contains three erroneous mentions of usage of a restriction enzyme— Bst Z17I—in the Methods section as displayed in the following sentences.
The original article was published in BMC Molecular Biology 2018 19 :3
Comparison of miRNA - 101a - 3p and miRNA - 144a - 3p regulation with the key genes of alpaca melanocyte pigmentation
Many miRNA functions have been revealed to date. Single miRNAs can participate in life processes by regulating more than one target gene, and more than one miRNA can also simultaneously act on one target mRNA....
Correction to: MicroRNA-325-3p protects the heart after myocardial infarction by inhibiting RIPK3 and programmed necrosis in mice
The original article [1] contains an error whereby Fig. 7 displays incorrect results; the correct version of Fig. 7 can be viewed ahead in this Correction article and should be considered in place of the origi...
The original article was published in BMC Molecular Biology 2019 20 :17
MicroRNA-325-3p protects the heart after myocardial infarction by inhibiting RIPK3 and programmed necrosis in mice
Receptor-interacting serine-threonine kinase 3 (RIPK3)-mediated necroptosis has been implicated in the progression of myocardial infarction (MI), but the underlying mechanisms, particularly whether microRNAs (...
The Correction to this article has been published in BMC Molecular Biology 2019 20 :18
Giant group I intron in a mitochondrial genome is removed by RNA back-splicing
The mitochondrial genomes of mushroom corals (Corallimorpharia) are remarkable for harboring two complex group I introns; ND5-717 and COI-884. How these autocatalytic RNA elements interfere with mitochondrial ...
Exploration of carbohydrate binding behavior and anti-proliferative activities of Arisaema tortuosum lectin
Lectins have come a long way from being identified as proteins that agglutinate cells to promising therapeutic agents in modern medicine. Through their specific binding property, they have proven to be anti-ca...
Characterization of cadmium-responsive MicroRNAs and their target genes in maize ( Zea mays ) roots
Current research has shown that microRNAs (miRNAs) play vital roles in plant response to stress caused by heavy metals such as aluminum, arsenic, cadmium (Cd), and mercury. Cd has become one of the most hazard...
Identification and validation of reference genes for real-time quantitative RT-PCR analysis in jute
With the availability of genome sequences, gene expression analysis of jute has drawn considerable attention for understanding the regulatory mechanisms of fiber development and improving fiber quality. Gene e...
Small nucleolar RNA Sf-15 regulates proliferation and apoptosis of Spodoptera frugiperda Sf9 cells
Small nucleolar RNAs (snoRNAs) function in guiding 2′- O -methylation and pseudouridylation of ribosomal RNAs (rRNAs) and small nuclear RNAs (snRNAs). In recent years, more and more snoRNAs have been found to play ...
Key genes differential expressions and pathway involved in salt and water-deprivation stresses for renal cortex in camel
Camels possess the characteristics of salt- and drought-resistances, due to the long-time adaption to the living environment in desert. The camel resistance research on transcriptome is rare and deficient, esp...
Development of a novel selection/counter-selection system for chromosomal gene integrations and deletions in lactic acid bacteria
The underlying mechanisms by which probiotic lactic acid bacteria (LAB) enhance the health of the consumer have not been fully elucidated. Verification of probiotic modes of action can be achieved by using sin...
Selection of reference genes for the quantitative real-time PCR normalization of gene expression in Isatis indigotica fortune
Isatis indigotica , a traditional Chinese medicine, produces a variety of active ingredients. However, little is known about the key genes and corresponding expression profiling involved in the biosynthesis pathwa...
MEF2A alters the proliferation, inflammation-related gene expression profiles and its silencing induces cellular senescence in human coronary endothelial cells
Myocyte enhancer factor 2A (MEF2A) plays an important role in cell proliferation, differentiation and survival. Functional deletion or mutation in MEF2A predisposes individuals to cardiovascular disease mainly...
Transcriptomic responses to grazing reveal the metabolic pathway leading to the biosynthesis of domoic acid and highlight different defense strategies in diatoms
A major cause of phytoplankton mortality is predation by zooplankton. Strategies to avoid grazers have probably played a major role in the evolution of phytoplankton and impacted bloom dynamics and trophic ene...
RNA sequencing, selection of reference genes and demonstration of feeding RNAi in Thrips tabaci (Lind.) (Thysanoptera: Thripidae)
Thrips tabaci is a severe pest of onion and cotton. Due to lack of information on its genome or transcriptome, not much is known about this insect at the molecular level. To initiate molecular studies in this ins...
A fragment activity assay reveals the key residues of TBC1D15 GTPase-activating protein (GAP) in Chiloscyllium plagiosum
GTPase-activating proteins (GAPs) with a TBC (Tre-2/Bub2/Cdc16) domain architecture serve as negative regulators of Rab GTPases. The related crystal structure has been studied and reported by other members of ...
HexA is required for growth, aflatoxin biosynthesis and virulence in Aspergillus flavus
Woronin bodies are fungal-specific organelles whose formation is derived from peroxisomes. The former are believed to be involved in the regulation of mycotoxins biosynthesis, but not in their damage repair fu...
Genome-wide identification of brain miRNAs in response to high-intensity intermittent swimming training in Rattus norvegicus by deep sequencing
Physical exercise can improve brain function by altering brain gene expression. The expression mechanisms underlying the brain’s response to exercise still remain unknown. miRNAs as vital regulators of gene ex...
Graphene oxide down-regulates genes of the oxidative phosphorylation complexes in a glioblastoma
Recently different forms of nanographene were proposed as the material with high anticancer potential. However, the mechanism of the suppressive activity of the graphene on cancer development remains unclear. ...
MiRNAs differentially expressed in skeletal muscle of animals with divergent estimated breeding values for beef tenderness
MicroRNAs (miRNAs) are small noncoding RNAs of approximately 22 nucleotides, highly conserved among species, which modulate gene expression by cleaving messenger RNA target or inhibiting translation. MiRNAs ar...
The Dictyostelium discoideum homologue of Twinkle, Twm1, is a mitochondrial DNA helicase, an active primase and promotes mitochondrial DNA replication
DNA replication requires contributions from various proteins, such as DNA helicases; in mitochondria Twinkle is important for maintaining and replicating mitochondrial DNA. Twinkle helicases are predicted to a...
Matrix association region/scaffold attachment region (MAR/SAR) sequence: its vital role in mediating chromosome breakages in nasopharyngeal epithelial cells via oxidative stress-induced apoptosis
Oxidative stress is known to be involved in most of the aetiological factors of nasopharyngeal carcinoma (NPC). Cells that are under oxidative stress may undergo apoptosis. We have previously demonstrated that...
Molecular analysis of NPAS3 functional domains and variants
NPAS3 encodes a transcription factor which has been associated with multiple human psychiatric and neurodevelopmental disorders. In mice, deletion of Npas3 was found to cause alterations in neurodevelopment, as w...
Integration of transcriptome and proteome profiles in glioblastoma: looking for the missing link
Glioblastoma (GB) is the most common and aggressive tumor of the brain. Genotype-based approaches and independent analyses of the transcriptome or the proteome have led to progress in understanding the underly...
Analyses of changes in myocardial long non-coding RNA and mRNA profiles after severe hemorrhagic shock and resuscitation via RNA sequencing in a rat model
Ischemia–reperfusion injury has been proven to induce organ dysfunction and death, although the mechanism is not fully understood. Long non-coding RNAs (lncRNAs) have drawn wide attention with their important ...
Coincidence cloning recovery of Brucella melitensis RNA from goat tissues: advancing the in vivo analysis of pathogen gene expression in brucellosis
Brucella melitensis bacteria cause persistent, intracellular infections in small ruminants as well as in humans, leading to significant morbidity and economic loss worldwide. The majority of experiments on the tr...
Positive cofactor 4 (PC4) contributes to the regulation of replication-dependent canonical histone gene expression
Core canonical histones are required in the S phase of the cell cycle to pack newly synthetized DNA, therefore the expression of their genes is highly activated during DNA replication. In mammalian cells, this...
Evaluation of suitable reference genes for qRT-PCR normalization in strawberry ( Fragaria × ananassa ) under different experimental conditions
Strawberry has received much attention due to its nutritional value, unique flavor, and attractive appearance. The availability of the whole genome sequence and multiple transcriptome databases allows the grea...
Laser capture microdissection for transcriptomic profiles in human skin biopsies
The acquisition of reliable tissue-specific RNA sequencing data from human skin biopsy represents a major advance in research. However, the complexity of the process of isolation of specific layers from fresh-...
Targeting miR-9 in gastric cancer cells using locked nucleic acid oligonucleotides
Gastric cancer is the third leading cause of cancer-related mortality worldwide. Recently, it has been demonstrated that gastric cancer cells display a specific miRNA expression profile, with increasing eviden...
Quantitative profiling of BATF family proteins/JUNB/IRF hetero-trimers using Spec-seq
BATF family transcription factors (BATF, BATF2 and BATF3) form hetero-trimers with JUNB and either IRF4 or IRF8 to regulate cell fate in T cells and dendritic cells in vivo. While each combination of the heter...
pH-mediated upregulation of AQP1 gene expression through the Spi-B transcription factor
Bicarbonate-based peritoneal dialysis (PD) fluids enhance the migratory capacity and damage-repair ability of human peritoneal mesothelial cells by upregulating AQP1. However, little is known about the underly...
A protocol for custom CRISPR Cas9 donor vector construction to truncate genes in mammalian cells using pcDNA3 backbone
Clustered regularly interspaced short palindromic repeat (CRISPR) RNA-guided adaptive immune systems are found in prokaryotes to defend cells from foreign DNA. CRISPR Cas9 systems have been modified and employ...
The Correction to this article has been published in BMC Molecular Biology 2019 20 :20
Recommendations for mRNA analysis of micro-dissected glomerular tufts from paraffin-embedded human kidney biopsy samples
Glomeruli are excellent pre-determined natural structures for laser micro-dissection. Compartment-specific glomerular gene expression analysis of formalin-fixed paraffin-embedded renal biopsies could improve r...
Nutrient depletion and TOR inhibition induce 18S and 25S ribosomal RNAs resistant to a 5′-phosphate-dependent exonuclease in Candida albicans and other yeasts
Messenger RNA (mRNA) represents a small percentage of RNAs in a cell, with ribosomal RNA (rRNA) making up the bulk of it. To isolate mRNA from eukaryotes, typically poly-A selection is carried out. Recently, a...
An optimized rapid bisulfite conversion method with high recovery of cell-free DNA
Methylation analysis of cell-free DNA is a encouraging tool for tumor diagnosis, monitoring and prognosis. Sensitivity of methylation analysis is a very important matter due to the tiny amounts of cell-free DN...
Sumoylation in p27kip1 via RanBP2 promotes cancer cell growth in cholangiocarcinoma cell line QBC939
Cholangiocarcinoma is one of the deadly disease with poor 5-year survival and poor response to conventional therapies. Previously, we found that p27kip1 nuclear-cytoplasmic translocation confers proliferation ...
An optimised protocol for isolation of RNA from small sections of laser-capture microdissected FFPE tissue amenable for next-generation sequencing
Formalin-fixed paraffin embedded (FFPE) tissue constitutes a vast treasury of samples for biomedical research. Thus far however, extraction of RNA from FFPE tissue has proved challenging due to chemical RNA–pr...
Physical shearing imparts biological activity to DNA and ability to transmit itself horizontally across species and kingdom boundaries
We have recently reported that cell-free DNA (cfDNA) fragments derived from dying cells that circulate in blood are biologically active molecules and can readily enter into healthy cells to activate DNA damage...
Interaction between NFATc2 and the transcription factor Sp1 in pancreatic carcinoma cells PaTu 8988t
Nuclear factors of activated T-cells (NFATs) have been mainly characterized in the context of immune response regulation because, as transcription factors, they have the ability to induce gene transcription. N...
Splicing arrays reveal novel RBM10 targets, including SMN2 pre-mRNA
RBM10 is an RNA binding protein involved in message stabilization and alternative splicing regulation. The objective of the research described herein was to identify novel targets of RBM10-regulated splicing. ...
Growth arrest specific gene 2 in tilapia ( Oreochromis niloticus ): molecular characterization and functional analysis under low-temperature stress
Growth arrest specific 2 ( gas2 ) gene is a component of the microfilament system that plays a major role in the cell cycle, regulation of microfilaments, and cell morphology during apoptotic processes. However, li...
Identification of G-quadruplex structures that possess transcriptional regulating functions in the Dele and Cdc6 CpG islands
G-quadruplex is a DNA secondary structure that has been shown to play an important role in biological systems. In a previous study, we identified 1998 G-quadruplex-forming sequences using a mouse CpG islands D...
Mitochondrial RNA processing in absence of tRNA punctuations in octocorals
Mitogenome diversity is staggering among early branching animals with respect to size, gene density, content and order, and number of tRNA genes, especially in cnidarians. This last point is of special interes...
Microarray expression profiling in the denervated hippocampus identifies long noncoding RNAs functionally involved in neurogenesis
The denervated hippocampus provides a proper microenvironment for the survival and neuronal differentiation of neural progenitors. While thousands of lncRNAs were identified, only a few lncRNAs that regulate n...
Early growth response protein 1 regulates promoter activity of α -plasma membrane calcium ATPase 2, a major calcium pump in the brain and auditory system
Along with sodium/calcium (Ca 2+ ) exchangers, plasma membrane Ca 2+ ATPases (ATP2Bs) are main regulators of intracellular Ca 2+ levels. There are four ATP2B paralogs encoded by four different genes. Atp2b2 encodes t...
BMC Molecular Biology
ISSN: 1471-2199
- General enquiries: [email protected]
Molecular Biology News
Top headlines, latest headlines.
- Pathogens Spread Unrecognized in the Body
- Enhanced Microscopy Analysis With AI
- Immune Response to Viruses in Mammal Cells
- Starving Cells Hijack Protein Transport Stations
- RNA's Hidden Potential: Future Bioengineering
- Algae Manipulates Microorganisms
- Solving a Mini Mystery of Cell Division
- Plant Sensors: Early Warning System for Farmers
- Tracking a Protein's Fleeting Shape Changes
- Human DNA Repair
Earlier Headlines
Monday, april 15, 2024.
- Unlocking the 'chain of Worms'
Friday, April 12, 2024
- Cell's 'garbage Disposal' May Have Another Role: Helping Neurons Near Skin Sense the Environment
- How Seaweed Became Multicellular
- Microbial Food as a Strategy Food Production of the Future
- Decoding the Language of Cells: Unveiling the Proteins Behind Cellular Organelle Communication
- Innovative Antiviral Defense With New CRISPR Tool
Thursday, April 11, 2024
- First Step to Untangle DNA: Supercoiled DNA Captures Gyrase Like a Lasso Ropes Cattle
- Genetic Underpinnings of Environmental Stress Identified in Model Plant
Wednesday, April 10, 2024
- The Genesis of Our Cellular Skeleton, Image by Image
- Researchers Discover How We Perceive Bitter Taste
- Cockayne Syndrome: New Insights Into Cellular DNA Repair Mechanism
Tuesday, April 9, 2024
- Machine Learning Method Reveals Chromosome Locations in Individual Cell Nucleus
Monday, April 8, 2024
- Different Means to the Same End: How a Worm Protects Its Chromosomes
- Bringing Multidrug-Resistant Pathogens to Their Knees
Friday, April 5, 2024
- Can Language Models Read the Genome? This One Decoded mRNA to Make Better Vaccines
Thursday, April 4, 2024
- Heat Flows the Secret to Order in Prebiotic Molecular Kitchen
Wednesday, April 3, 2024
- New Tools Reveal How Genes Work and Cells Organize
- Evolution in Action? New Study Finds Possibility of Nitrogen-Fixing Organelles
- New Discovery Unravels Malaria Invasion Mechanism
- First View of Centromere Variation and Evolution
Friday, March 29, 2024
- Connecting the Dots to Shape Growth Forces
Thursday, March 28, 2024
- How the Crimean-Congo Hemorrhagic Fever Virus Enters Our Cells
- Researchers Discover Key Gene for Toxic Alkaloid in Barley
- Cell Division Quality Control 'stopwatch' Uncovered
Wednesday, March 27, 2024
- New Technique for Predicting Protein Dynamics May Prove Big Breakthrough for Drug Discovery
Tuesday, March 26, 2024
- Researchers Show That Introduced Tardigrade Proteins Can Slow Metabolism in Human Cells
- Discovery of Amino Acid Unveils How Light Makes Plants Open
Friday, March 22, 2024
- Natural Recycling at the Origin of Life
Thursday, March 21, 2024
- As We Age, Our Cells Are Less Likely to Express Longer Genes
- Natural Molecule Found in Coffee and Human Body Increases NAD+ Levels, Improves Muscle Function During Aging
- Decoding the Plant World's Complex Biochemical Communication Networks
Tuesday, March 19, 2024
- A Protein Found in Human Sweat May Protect Against Lyme Disease
Monday, March 18, 2024
- How Cells Are Ahead of the Curve
- Engineers Measure pH in Cell Condensates
Friday, March 15, 2024
- Machine Learning Classifier Accelerates the Development of Cellular Immunotherapies
Thursday, March 14, 2024
- Small Amounts of Licorice Raise Blood Pressure, Study Finds
- Alzheimer's Drug Fermented With Help from AI and Bacteria Moves Closer to Reality
- Sleep-Wake Rhythm: Fish Change Our Understanding of Sleep Regulation
- New Discovery Reveals How the Egg Controls Sperm Entry
- Even Cells Know the Importance of Recycling
Wednesday, March 13, 2024
- Sulfur and the Origin of Life
- Tryptophan in Diet, Gut Bacteria Protect Against E. Coli Infection
- New Computational Strategy Boosts the Ability of Drug Designers to Target Proteins Inside the Membrane
- Milk to the Rescue for Diabetics? Cow Produces Human Insulin in Milk
Tuesday, March 12, 2024
- A Simple and Robust Experimental Process for Protein Engineering
- Scientists Find Weak Points on Epstein-Barr Virus
- How a Natural Compound from Sea Squirts Combats Cancer
Monday, March 11, 2024
- Researchers Uncover Protein Responsible for Cold Sensation
Friday, March 8, 2024
- Research Sheds Light on New Strategy to Treat Infertility
- New Study Discovers How Altered Protein Folding Drives Multicellular Evolution
- Researchers Develop Artificial Building Blocks of Life
- Mutation Solves a Century-Old Mystery in Meiosis
Thursday, March 7, 2024
- Vitamin A May Play a Central Role in Stem Cell Biology and Wound Repair
- The Malaria Parasite Generates Genetic Diversity Using an Evolutionary 'copy-Paste' Tactic
- Cracking Epigenetic Inheritance: Biologists Discovered the Secrets of How Gene Traits Are Passed on
Wednesday, March 6, 2024
- First Heat Map for Individual Red Blood Cells
- Universal Tool for Tracking Cell-to-Cell Interactions
- Synthetic Gene Helps Explain the Mysteries of Transcription Across Species
- Decoding the Language of Epigenetic Modifications
- Deconstructing the Structural Elements of a Lesser-Known Microbe
Tuesday, March 5, 2024
- Microalgae With Unusual Cell Biology
Monday, March 4, 2024
- Modeling the Origins of Life: New Evidence for an 'RNA World'
- Photosynthetic Secrets Come to Light
Friday, March 1, 2024
- Light Into the Darkness of Photosynthesis
- How Virus Causes Cancer: Potential Treatment
- Convergent Evolution of Algal CO2-Fixing Organelles
Thursday, February 29, 2024
- New Role for Bacterial Enzyme in Gut Metabolism Revealed
- Scientists Identify New 'regulatory' Function of Learning and Memory Gene Common to All Mammalian Brain Cells
- Scientists Develop Novel RNA Or DNA-Based Substances to Protect Plants from Viruses
- The Golgi Organelle's Ribbon Structure Is Not Exclusive to Vertebrates, Contrary to Previous Consensus
- How First Cells Could Have Formed on Earth
- Radio Waves Can Tune Up Bacteria to Become Life-Saving Medicines
Wednesday, February 28, 2024
- New Tool Helps Decipher Gene Behavior
- Nanocarrier With Escape Reflex
- Change in Gene Code May Explain How Human Ancestors Lost Tails
Tuesday, February 27, 2024
- New Discovery Shows How Cells Defend Themselves During Stressful Situations
- Scientists Use Blue-Green Algae as a Surrogate Mother for 'meat-Like' Proteins
- Microbial Comics: RNA as a Common Language, Presented in Extracellular Speech-Bubbles
- Low-Temperature Plasma Used to Remove E. Coli from Hydroponically Grown Crops
Monday, February 26, 2024
- Human Stem Cells Coaxed to Mimic the Very Early Central Nervous System
- Cutting-Edge 'protein Lawnmower' Created
- The Small Intestine Adapt Its Size According to Nutrient Intake
Friday, February 23, 2024
- Biomolecular Condensates -- Regulatory Hubs for Plant Iron Supply
- Ribosomes: Molecular Wedge Assists Recycling
Thursday, February 22, 2024
- Compound Vital for All Life Likely Played a Role in Life's Origin
- Metabolic Diseases May Be Driven by Gut Microbiome, Loss of Ovarian Hormones
- Damage to Cell Membranes Causes Cell Aging
Wednesday, February 21, 2024
- New System Triggers Cellular Waste Disposal
Tuesday, February 20, 2024
- Researchers Are Using RNA in a New Approach to Fight HIV
- Bridging Diet, Microbes, and Metabolism: Implications for Metabolic Disorders
- Photosynthetic Mechanism of Purple Sulfur Bacterium Adapted to Low-Calcium Environments
Monday, February 19, 2024
- Eating Too Much Protein Is Bad for Your Arteries, and This Amino Acid Is to Blame
Friday, February 16, 2024
- Toxoplasmosis: Evolution of Infection Machinery
Thursday, February 15, 2024
- Asexual Propagation of Crop Plants Gets Closer
- Ancient Retroviruses Played a Key Role in the Evolution of Vertebrate Brains
Wednesday, February 14, 2024
- Team Creates Novel Rabies Viral Vectors for Neural Circuit Mapping
- Key Genes Linked to DNA Damage and Human Disease Uncovered
- Researchers Uncover Mechanisms Behind Enigmatic Shapes of Nuclei
- By Growing Animal Cells in Rice Grains, Scientists Dish Up Hybrid Food
- Why Do Flies Fall in Love? Researchers Tease out the Signals Behind Fruit Fly Courtship Songs
- LATEST NEWS
- Top Science
- Top Physical/Tech
- Top Environment
- Top Society/Education
- Health & Medicine
- Mind & Brain
- Living Well
- Space & Time
- Matter & Energy
- Computers & Math
- Plants & Animals
- Agriculture & Food
- Beer and Wine
- Bird Flu Research
- Genetically Modified
- Pests and Parasites
- Cows, Sheep, Pigs
- Dolphins and Whales
- Frogs and Reptiles
- Insects (including Butterflies)
- New Species
- Spiders and Ticks
- Veterinary Medicine
- Business & Industry
- Biotechnology and Bioengineering
- CRISPR Gene Editing
- Food and Agriculture
- Endangered Animals
- Endangered Plants
- Extreme Survival
- Invasive Species
- Wild Animals
- Education & Learning
- Animal Learning and Intelligence
- Life Sciences
- Behavioral Science
- Biochemistry Research
- Biotechnology
- Cell Biology
- Developmental Biology
- Epigenetics Research
- Evolutionary Biology
- Marine Biology
- Mating and Breeding
- Molecular Biology
- Microbes and More
- Microbiology
- Zika Virus Research
- Earth & Climate
- Fossils & Ruins
- Science & Society
Strange & Offbeat
- Fossil Frogs Share Their Skincare Secrets
- Fussy Eater? Most Parents Play Short Order Cook
- Precise Time Measurement: Superradiant Atoms
- Artificial Cells That Act Like Living Cells
- Affordable and Targeted Anticancer Agent
- This Alloy Is Kinky
- Giant Galactic Explosion: Galaxy Pollution
- Flare Erupting Around a Black Hole
- Two Species Interbreeding Created New Butterfly
- Warming Antarctic Deep-Sea and Sea Level Rise
Trending Topics
When you choose to publish with PLOS, your research makes an impact. Make your work accessible to all, without restrictions, and accelerate scientific discovery with options like preprints and published peer review that make your work more Open.
- PLOS Biology
- PLOS Climate
- PLOS Complex Systems
- PLOS Computational Biology
- PLOS Digital Health
- PLOS Genetics
- PLOS Global Public Health
- PLOS Medicine
- PLOS Mental Health
- PLOS Neglected Tropical Diseases
- PLOS Pathogens
- PLOS Sustainability and Transformation
- PLOS Collections
Molecular Biology
Empowering a community publishing articles in Molecular Biology, including RNA biology, chromatin and epigenetics, gene expression, translation, DNA replication and repair, nuclear organization, gene editing, and much more.
At PLOS, we put researchers and research first.
Our expert editorial boards collaborate with reviewers to provide accurate assessment that readers can trust. Authors have a choice of journals, publishing outputs, and tools to open their science to new audiences and get credit. We collaborate to make science, and the process of publishing science, fair, equitable, and accessible for the whole community.
PLOS publishes a suite of influential Open Access journals across all areas of science and medicine.
Rigorously reported, peer reviewed and immediately available without restrictions, promoting the widest readership and impact possible. We encourage you to consider the journal’s scope before submission, as they are all editorially independent and specialized in their publication criteria and breadth of content.
Looking for exciting work in your field?
Discover top cited Molecular Biology papers from recent years.
PLOS ONE Glyoxal fixation facilitates transcriptome analysis after antigen staining and cell sorting by flow cytometry Read the preprint...
PLOS PATHOGENS Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2 Read the peer review...
Discover a range of antimicrobial resistance research compiled by the expert guest editors from PLOS Medicine , PLOS Biology and PLOS ONE .
Reproducibility is important for the future of science.
PLOS is Open so that everyone can read, share, and reuse the research we publish. Underlying our commitment to Open Science is our data availability policy which ensures every piece of your research is accessible and replicable. We also go beyond that, empowering authors to preregister their research, and publish protocols , negative and null results, and more.

With trainig in molecular biology, Ines joined PLOS Biology in 2008 and enjoys publishing stories that advance science. Learn about her career as an editor and advice for early career researchers new to publishing.

How can we increase adoption of open research practices?

Researchers are satisfied with their ability to share their own research data but may struggle with accessing other researchers’ data. Therefore, to increase data sharing in a findable and accessible way, PLOS will focus on better integrating existing data repositories and promoting their benefits rather than creating new solutions.
Imagining a transformed scientific publication landscape

Open Science is not a finish line, but rather a means to an end. An underlying goal behind the movement towards Open Science is to conduct and publish more reliable and thoroughly reported research.
Editors' picks 2020

Here, PLOS ONE Staff Editors from the different subject teams reflect on the past year choosing some of their favorite research. From research on plastic pollution to improving prognosis predictions for patients with cancer, we hope that these selections will have something of interest for everyone.
- What do you think is the best way to ensure reproducibility for future generations of researchers?
BMC Molecular and Cell Biology
Collections open for submissions.
Cell size and cell division mechanics
Guest edited by Shane McInally and Kristi Miller
Organoids as models of development and disease
Guest edited by Sérgio Paulo Bydlowski
From germ cells to fertilization: understanding the development of gametes
Guest edited by Su-Ren Chen
Responding to stress: cellular and molecular mechanisms
Guest edited by Francesco Cappello and Ryo Higuchi-Sanabria
Editor's Choice
Keratin 19 binds and regulates cytoplasmic HNRNPK mRNA targets in triple-negative breast cancer
Genetic and protein interaction studies between the ciliary dyslexia candidate genes DYX1C1 and DCDC2
Myogenic differentiation of human myoblasts and Mesenchymal stromal cells under GDF11 on Poly-ɛ-caprolactone-collagen I-Polyethylene-nanofibers
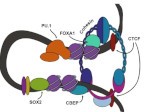
Chromatin structure in cancer
In this new Review Wang et al. describe recent insights into altered genome architecture in human cancer, highlighting multiple pathways that disrupt chromatin structure and contribute to tumorgenesis.
- Most accessed
mTOR signaling pathway regulation HIF-1 α effects on LPS induced intestinal mucosal epithelial model damage
Authors: Zeyong Huang, Wenbin Teng, Liuxu Yao, Kai Xie, Suqin Hang, Rui He and Yuhong Li
Long non-coding RNA SOX2OT in tamoxifen-resistant breast cancer
Authors: Jeeyeon Lee, Eun-Ae Kim, Jieun Kang, Yee Soo Chae, Ho Yong Park, Byeongju Kang, Soo Jung Lee, In Hee Lee, Ji-Young Park, Nora Jee-young Park and Jin Hyang Jung
Mice lacking DIO3 exhibit sex-specific alterations in circadian patterns of corticosterone and gene expression in metabolic tissues
Authors: Zhaofei Wu, M. Elena Martinez and Arturo Hernandez
Optimization of seeding density of OP9 cells to improve hematopoietic differentiation efficiency
Authors: Xin-xing Jiang, Meng-yi Song, Qi Li, Yun-jian Wei, Yuan-hua Huang and Yan-lin Ma
Development of an in vitro human alveolar epithelial air-liquid interface model using a small molecule inhibitor cocktail
Authors: Ikuya Tanabe, Kanae Ishimori and Shinkichi Ishikawa
Most recent articles RSS
View all articles
Hypoxia-mimetic agents inhibit proliferation and alter the morphology of human umbilical cord-derived mesenchymal stem cells
Authors: Hui-Lan Zeng, Qi Zhong, Yong-Liang Qin, Qian-Qian Bu, Xin-Ai Han, Hai-Tao Jia and Hong-Wei Liu
Multiple immunofluorescence labelling of formalin-fixed paraffin-embedded (FFPE) tissue
Authors: David Robertson, Kay Savage, Jorge S Reis-Filho and Clare M Isacke
Caspase-9, caspase-3 and caspase-7 have distinct roles during intrinsic apoptosis
Authors: Matthew Brentnall, Luis Rodriguez-Menocal, Rebeka Ladron De Guevara, Enrique Cepero and Lawrence H Boise
Interleukin-1α enhances the aggressive behavior of pancreatic cancer cells by regulating the α 6 β 1 -integrin and urokinase plasminogen activator receptor expression
Authors: Hirozumi Sawai, Yuji Okada, Hitoshi Funahashi, Yoichi Matsuo, Hiroki Takahashi, Hiromitsu Takeyama and Tadao Manabe
Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence
Authors: Eva Latorre, Vishal C. Birar, Angela N. Sheerin, J. Charles C. Jeynes, Amy Hooper, Helen R. Dawe, David Melzer, Lynne S. Cox, Richard G. A. Faragher, Elizabeth L. Ostler and Lorna W. Harries
Most accessed articles RSS
Aims and scope
BMC Molecular and Cell Biology , formerly known as BMC Cell Biology , is an open access journal that considers articles on all aspects of cellular and molecular biology in both eukaryotic and prokaryotic cells. The journal considers studies on functional cell biology, molecular mechanisms of transcription and translation, biochemistry, as well as research using both the experimental and theoretical aspects of physics to study biological processes and investigations into the structure of biological macromolecules.

We are recruiting!
We are currently looking for new Editorial Board Members to join our team of academic editors, assessing manuscripts in the field of molecular and cell biology.
BMC Series Blog

Introducing BMC Primary Care’s Collection: Trust and mistrust in primary care
17 April 2024

Highlights of the BMC Series – March 2024
10 April 2024

Introducing BMC Bioinformatics’ Collection: Bioinformatics ethics and data privacy
09 April 2024
Latest Tweets
Your browser needs to have JavaScript enabled to view this timeline
Important information
Editorial board
For authors
For editorial board members
For reviewers
- Manuscript editing services
Annual Journal Metrics
2022 Citation Impact 2.8 - 2-year Impact Factor 2.9 - 5-year Impact Factor 0.678 - SNIP (Source Normalized Impact per Paper) 0.775 - SJR (SCImago Journal Rank)
2023 Speed 24 days submission to first editorial decision for all manuscripts (Median) 143 days submission to accept (Median)
2023 Usage 582,382 downloads 193 Altmetric mentions
- More about our metrics
- Follow us on Twitter
ISSN: 2661-8850
- General enquiries: [email protected]
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List
- Iran J Public Health
- v.46(11); 2017 Nov
Role of Molecular Biology in Cancer Treatment: A Review Article
1. Center of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan
Hafiza Yasara QAMAR
2. Institute of Molecular Biology and Biotechnology, University of Lahore, Lahore, Pakistan
Hafsa NAEEM
Mariam riaz, naila kanwal.
3. Dept. of Plant Breeding and Genetics, University of Agriculture, Faisalabad, Pakistan
4. Southern Cross Plant Science, Southern Cross University, Lismore, Australia
Muhammad Farooq SABAR *
5. Centre for Applied Molecular Biology, University of the Punjab, Lahore, Pakistan
Idrees Ahmad NASIR
Background:.
Cancer is a genetic disease and mainly arises due to a number of reasons include activation of onco-genes, malfunction of tumor suppressor genes or mutagenesis due to external factors.
This article was written from the data collected from PubMed, Nature, Science Direct, Springer and Elsevier groups of journals.
Oncogenes are deregulated form of normal proto-oncogenes required for cell division, differentiation and regulation. The conversion of proto-oncogene to oncogene is caused due to translocation, rearrangement of chromosomes or mutation in gene due to addition, deletion, duplication or viral infection. These oncogenes are targeted by drugs or RNAi system to prevent proliferation of cancerous cells. There have been developed different techniques of molecular biology used to diagnose and treat cancer, including retroviral therapy, silencing of oncogenes and mutations in tumor suppressor genes.
Conclusion:
Among all the techniques used, RNAi, zinc finger nucleases and CRISPR hold a brighter future towards creating a Cancer Free World.
Introduction
Cancer is a genetic disease. The expression of oncogenesis is an important event in early stages of tumor formation. Oncogenes are activated through two mechanisms: either by infection of cells by tumor viruses or by mutation of cellular proto-oncogenes (which are usually normal) to oncogenes. Then tumors originate by oncogenic transformation of only a single cell. Some tumors adopt the ability to escape the site of their origin and intrude other parts of the body. This process is called metastasis. Solid tumors i.e. sarcomas, could be transferred from one animal to another using Rous sarcoma virus. Tumors could be caused either by the addition or by expression of genetic material, which in this case was viral DNA, to normal cells. Rous was presented a Nobel Prize for his work. In 1978, tumors of nonviral origin were also discovered ( 1 ).
This article was written from the data collected from well-reputed `databases of PubMed, Science Direct, Springer and Elsevier groups of journals. The data were collected in the form of research and review articles on the use of molecular biology for cancer research, which was summarized to form an improved and advance review article. The following key words were used to search the databases: Cancer, Oncogenes, Proto-oncogenes, Mutagenesis, Viral infection, Tumor, CRISPR. Articles were verified published until 2015.
Results and discussion
Further, the relation between carcinogens and mutagens saying was described that many carcinogens are also mutagens. Mutations in normal genes might cause them to become cancerous and this could happen due to any damage to DNA. This hypothesis got further support after molecular biology tools were applied in cancer research ( 2 ). Other than expression of oncogenes, cancer may also arise due to the loss or damage of tumor suppressor genes that check and control the cell growth. In many cases, both reasons contribute to causing cancer ( 3 ).

Oncogenes in cancer development
Oncogenes are the tumor causing genes and have important role in development of many cancers. In 1970, SRC oncogene was discovered in chicken retrovirus. As a result of some mutation in the otherwise normal proto-oncogenes, their deregulation occurs and uncontrolled proliferation of cells starts and leads to cancer ( 4 ). At genomic level, only single oncogenic allele is required to alter normal gene function because of its dominant property. The origin of oncogene can be cellular i.e., from inside the body or viral i.e., from some virus ( 3 ).
Gene Duplication, addition, insertion, deletion or chromosomal translocation, chromosomal rearrangement of certain proto-oncogenes alters their function and converts them into oncogenes. These mutations overexpress the protein to an uncontrolled level, which may lead to tumor. These mutations may occur due to external factors or internal factors or both like viral infection, radiation or chemicals, injury and disease ( 4 ). Among these mutations, viral infection is the rare cause of oncogene activation in animals but is of great importance for understanding oncogene function.
Viral infection
Retroviruses or DNA viruses cause viral infections. These viruses infect the host either by inserting oncogenes in host chromosome, interfering proto-oncogene transcription factors/regulators or by inserting homologous sequences corresponding to normal protooncogene of host. For example, retrovirus carrying SRC oncogenes infects the host, integrates viral chromosome in host chromosome, further divides the viral progeny and infects the surrounding cells, inducing overexpression of cellular normal genes and deregulated proliferation of cells in order to cause cancer ( 5 ).
Types and classification of oncogenes:
Oncogenes can be classified into five classes based on protein products formed by mutation or deregulation of proto-oncogenes. These include growth factors, growth hormone/factor receptors (GRFs), serine/threonine kinases, GTPase molecules and, transcription factors. Mutations in the growth factors can lead to several types of cancers such as fibrosarcoma, glioblastoma (brain cancer), osteosarcoma (bone cancer) etc. ( 6 , 7 ).
In several tumors, “ligand-binding domain” deletions of Epidermal Growth Factor Receptor cause successive activation of receptor even in absence of ligand by transmembrane protein carrying tyrosine-kinase activity. This activation causes interaction with further cytoplasmic proteins like “SRC domain” and leads to deregulation of numerous signaling pathways. Mostly in gastrointestinal, breast and lung cancers, GFR mutations occur ( 4 ). Similarly, overexpression of Raf-1 kinase and cyclin-dependent kinases due to uncontrolled phosphorylation may cause many cancers such as thyroid and ovarian cancer.
Deregulated activation of GTPases such as Ras, causes activation of MAPK pathway and uncontrolled signaling and division of cells cause several cancers such as myeloid leukemia. Transcription factor proteins are products of protooncogenes. The mutation, translocation or rearrangement of these causes overexpression of gene and unwanted consecutive transcription of target gene that leads to any types of cancers such as pancreatic and lung cancer ( 6 ).
Role of oncogenes to treat cancer
The oncogenes are targeted to treat oncogenic cancer. Several oncogenes discussed above are targeted by drugs and gene therapies to inhibit, arrest, regulate or senescence their genes. For example, Imatinib (ABL kinase inhibitor) or Gleevec is used to treat BCR-ABL. Gefitinib or Iressa, erlotinib or Tarceva are used to target EGFR. VEGF oncogenes are targeted by bevacizumab or sorafenib. Sorafenib is also used to downregulate or inhibit B-Raf oncogene. These agents/drugs are used, sometimes in combination, for chemotherapy to inhibit proliferation of oncogenes or to downregulate signaling oncoproteins in several signaling pathways to treat oncogenic cancers ( 8 ). However, it is difficult to target “non-kinase oncogenes” through drugs such as Myc and Ras.
Tumor suppressor genes in cancer
Tumor suppressors play their role by inhibiting cellular proliferation and tumor development. In most of the tumors, inactivation of the tumor suppressor genes eliminates the negative regulation of these genes over cellular proliferation that leads to abnormal cell proliferation, therefore, causing cancer. Tumor suppressor genes have “loss of function” mutations because they develop cancer by inactivating their inhibitory effect on cell proliferation. For a tumor suppressor gene to promote tumor development, both copies of the gene must be inactivated because one copy is sufficient for controlling cell proliferation. These mutations act recessively ( 9 ).
Role of Tumor Suppressor Genes
The protein products tumor suppressing genes are found to play the following important functions:
- Enzymes involved in DNA repair
- Checkpoint-control proteins arresting the cell cycle in case DNA is impaired or chromosome abnormality.
- Proteins promoting programmed cell death (apoptosis)
- Inhibiting cellular growth and proliferation by acting as receptors for hormones.
- Intracellular proteins which regulate or inhibits movement through a specific stage of cell cycle ( 10 ).
Role of Tumor Suppressor Genes in Cancer Wilms’ Tumor 1 Gene
Some tumor-suppressing genes act as transcriptional regulatory proteins. For example, the product of WT1 gene which is a repressor protein and acts by suppressing transcription of many growth factor-inducible genes. WT1 is made inactive in Wilms’ tumors (which is a tumor in kidney found in children). Insulin-like growth factor II is the target of WT1 gene, over-expressed in Wilms’ tumors, thereby contributing to abnormal cell proliferation ( 11 ).
Retinoblastoma and INK4 Genes
Several tumor suppressor genes regulate cell cycle progression through a specific stage e.g. protein products of Rb and INK-4 genes. Retinoblastoma is the tumor of the eye. Two mutagenic events are required for the retinoblastoma development in sporadic cases whereas only one mutagenic event is needed in individuals with inherited form of the disease in which it displays autosomal dominant inheritance. In normal cells, Cdk2 and cyclin D complexes regulate the entry through the constraint point, thereby phosphorylating and inactivating pRb. pRb also impedes the entry through the constraint point in the G 1 phase of the cell cycle by repressing the transcription of many genes involved in cell cycle advancement. The INK4 tumor suppressor gene also regulates movement through the restriction point by encoding Cdk inhibitor p16. Inactivation of INK4 results in uncontrolled phosphorylation of Rb ( 10 ).
p53 Tumor Suppressor Gene
The p53 plays its role by regulating cell cycle and programmed cell death. The p53 can arrest the cell cycle upon DNA damage. It allows the DNA to repair or cause the programmed cell death (apoptosis). This is achieved by activating a number of genes involved in controlling and regulating the cell cycle. Mutation in p53 in tumorigenic cells results in uncontrolled cell proliferation and inefficient DNA repair. p53 mutations are estimated to be the most common in tumors of humans, approximately 50% or even greater than that ( 12 ).
Breast Cancer-1 and 2 Genes
Breast cancer-1 and 2 genes are linked to familial breast cancer. Breast Cancer-1 gene consists of 100 Kb DNA and 21 exons. It has a zinc-finger domain like that in the DNA binding proteins. Breast cancer-1 is a tumor suppressor gene. BRCA-2 is located on chromosome 13.
Tumor Suppressor Genes and their application
Tumor suppressor genes can be studied at the levels of DNA, mRNA, and proteins in the normal and cancerous cells using various methods. Tests for the detection of heterozygosity can be helpful for identifying individuals predisposed to retinoblastoma and other malignancies. Higher frequency of p53 mutations also offers diagnostic and analytical possibilities. PCR amplification can be used to study the changes along with recent methods such as RNase protection assays, single-strand conformational polymorphism or denaturing gel electrophoresis. Immunometric–type assays are quite good at measuring altered p53 in tumor cell line lysates and tissue homogenates ( 13 , 14 ).
Molecular pathology: Diagnosis of cancer
One of the primary challenges in the clinical management of cancer patients is to establish the correct diagnosis. As a result, a number of technologies have been developed and are now routinely employed to subtype molecularly cancers. These include immunohistochemistry, immunofluorescence, and the analysis of DNA and RNA extracted from the lesion-through In situ hybridization and fluorescent in situ hybridization (FISH). The cancer specimen is then subtyped using different approaches of molecular biology including Sanger sequencing, pyrosequencing, allele-specific PCR. Cancer genotyping is performed by snapshot assay, mass spectroscopy based assays and next generation sequencing (based on fluorescence or semiconductor detectors). The introduction of next-generation sequencing is serving to uncover the true diversity of cancers as well as to define recurring mutations targeted with new therapies. Such genomic-level analyses will continue to have an impact for many years ( 15 , 16 ).
Cancer treatment – then & now
Different treatment techniques and therapies have been applied for the treatment of cancer at different times. Some of the most common methods used include surgery, radiation therapy, chemotherapy, hormonal therapy, immunotherapy, adjuvant therapy, targeted-growth signal inhibition, drugs that induce apoptosis, nanotechnology, RNA expression and profiling, and the latest being CRISPR ( 17 ). Few of these shall be later discussed in this review.
Cancer cells can also be killed by gene replacements or by knocking out oncogenes. Oncolytic viruses can be used in combination with chemo-therapeutic agents to destroy cancer cells as well ( 18 ).
Retroviral therapy for cancer
Apart from the conventional methods, retroviruses (RVs) have also been used in cancer therapy. RVs can be and have been used for transferring genes to mammalian cells. Most popular RVs are the ones derived from the Moloney Murine leukaemia virus (MoMLV). In the last twenty years, artificial evolution of RVs has enabled their applications in developing transgenic animals, stable delivery of siRNA and clinical trials for gene therapy. The great potential of RVs was discussed in recent reports about the successful clinical trials of gene therapy in patients with severe immunodeficiency disease. However, there are some probable risks inherently related with that ( 19 ). A comparative experiment was done by designing two groups of vectors. One was intrinsically replicative, the other was defective and so it had a helper retrovirus with it. Under in vitro conditions, the replicative viruses achieved more than 85% transduction while the other transduced only less than 1%. This experiment clearly indicates the potential of RRVs for developing cancer gene therapy ( 20 ).
Retroviral tagging and insertional oncogenesis
Recently, interest in investigating retroviral vector insertions (insertional oncogenesis) has been growing. For many years, viral insertion sites were used to identify possible oncogenes and cancer signaling pathways. The scope of this approach has been broadened by techniques like insertion site cloning by high throughput PCR, availability of genetically modified animals and with the completion of mouse genome project ( 21 , 22 ). However, numerous investigators have identified hundreds of common sites. These integration sites are usually associated with cancer genes in MoMLV-induced murine haematopoietic malignancies ( 23 – 25 ). Generally, majority of the insertions exist outside the coding regions. Therefore, only less than 10% of the RISs can be considered as the accepted tumor suppressor genes. Quite interestingly, around 17%–18% of RISs are targeted transcription factors ( 19 ).
Problems with Retroviral Therapy
There are some safety concerns associated with the retroviral gene therapy addressed. Some possible solutions to this problem could be targeted infection, transcriptional targeting, local delivery, co-transduction of retroviral vector with a suicidal gene, specifically targeted retroviral insertion, and SIN vectors. These procedures, with both viral and non-viral systems, can be used in protocols for gene therapy. Nevertheless, insertional oncogenesis remains the major concern regarding retroviral gene therapy ( 26 ). A safe and fast means of targeted approach could be the use of foamy viruses ( 27 , 28 ). These viruses are harmless to humans and yet have a wide range of hosts ( 29 – 31 ). However, for ex vivo cell-based gene therapy, insertional oncogenesis can be avoided by pre-screening of transduced cells in order to select only those cells (clones) which have the transgene only at a desirable site of the chromosome ( 26 ).
Molecular biology techniques for the treatment of cancer
Initially, homologous recombination was used to inactivate the target gene. This was done for characterizing the function of genes. This method was not as productive as it was not efficient in introducing constructs onto the target site. It was very lengthy process, the selection process was laborious and there were many severe mutagenic effects of this method ( 32 ).
RNA Interference
One of the methods used in cancer therapy is RNA interference. RNA interference involves the use of small non-coding RNA that can bind with other mRNAs and can inhibit their translation into proteins. This can result in the loss of function of genes. In cancer therapy, these RNAi can be used to destroy the function of cancer genes that prevent cancer from spreading ( 33 – 35 ). The RNAi method is fast, cheap and it has a high efficiency so it replaced homologous recombination method. Still, it has drawbacks that include incomplete knockdown and temporary prevention of gene function. It also gives off-target effects unpredicted. Cancer recurrence was also seen in some cases. All these problems lead the researchers to explore new methods to alter the gene function ( 36 ).
Genome editing is a far better and new technique to treat cancer. Scientists used engineered nucleases that have specific domains that can bind to the target site followed by its cleavage ( 37 , 38 ). These nucleases were able to induce double-strand breaks (DSBs) in the target followed by the activation of DNA repair mechanisms. Two types of nucleases are used that include programmable nucleases like Zinc Finger Nucleases (ZFNs) and transcription-activator-like effector nucleases (TALENs). These nucleases were successful in genome editing for curing cancer in different animal models ( 39 ).
Zinc finger proteins (ZNFs)
Znfs are the first nucleases to be used for gene editing.
These are found as DNA binding domains in eukaryotes. These are made up of 30 amino acids modules arranged in the form of an array of Cys2-His2 DNA-binding zinc fingers. These modules are used to a nuclease domain of FokI ( 37 , 40 , 41 ). The modules consist of 3–6 zinc fingers that can identify nucleotide triplets ( 42 , 43 ). The FokI nuclease functions only as dimers so a pair of zinc finger nucleases is needed to target any region in the genome. One ZFN will identify the sequence upstream of the genome region to be modified and other will identify downstream sequence ( 44 ). These arrays bind to nearby DNA sequences that are in the opposite strands to induce a double-stranded break in the specific region. The breaks are then repaired by different methods that can cause different changes in the specific region like point mutations, indels or translocations. The ZNFs are custom designed so that they recognize all possible nucleotides and any specific region of DNA. ( 42 , 45 , 46 ).
Transcription activator-like effector nucleases (TALENs)
TALENs are similar to zinc finger nucleases, as they also need DNA binding motifs and the same nuclease to edit the genome. The difference lies in the recognition of nucleotides as the TALENs domain identifies only one nucleotide instead of a triplet. The interactions between the TALEN domains and their target sites are stronger as compared to ZNFs. It is easier to design TALENs as compared to designing of ZNFs ( 46 , 47 ). Using TALENs for cancer therapy is very effective method. It needs two specifically engineered TALENs that can identify the sequences of DNA in the target gene on the opposite strands. It dimerizes the FokI nuclease cleavage domain in the TALENs, cleaving the sequence in the target gene ( 48 – 50 ). This causes double-stranded DNA breaks in the targeted gene. The lesion due to the DNA break is repaired by the end-joining DNA repair system. The target gene is altered due to the change in the reading frame. This method can also be used to remove the already present mutations. This gene editing technology can be used to treat the cancer cell lines efficiently as it can target any gene in the genome. Complex cancer genes can be treated by using TALENs ( 51 ).
CRISPR/CAS9 system: A powerful tool for genome editing in cancer
A powerful genome-editing technology known as Clustered regularly interspaced palindromic sequences-acronym CRISPR, is now eclipsing all other genome-engineering techniques. This revolutionary technique allows researchers to accomplish targeted manipulation in any gene (DNA sequence) in the entire genome of any organism in vitro or now even directly in endogenous genome, thus helping to elucidate the functional organization of genome at systems level and identifying casual genetic variations. CRISPR plays a vital role in detection of cancers ( 52 ).
Mechanism of CRISPR-Cas9 in cancer treatment
If cancer causing gene is known, cancerous cells can be treated with CRISPR-Cas9 system which helps in gene deletion and replacement with a normal gene. Yin et al in his paper discuss the injection process using CRISPR-Cas9 system to cut and introduce gene into liver cells. CRISPRCas9 is more effective for single gene mutation cancers and is mostly delivered in vitro in a particular location. In case of metastatic cancers, in vitro delivery becomes difficult. CRISPR primarily constitutes two biological components: Engineered single guide RNA (sgRNA) and Cas9. A small guide RNA (crRNA and tracrRNA) is used to recognize the complementary sequence-specific target flanked by proto-spacer adjacent motif (PAM) and it guides endonucleases i.e. Cas9 to cleave this sequence ( 53 ). Different CRISPR-Cas systems have been grouped majorly into three types (I–III) and subtypes (as I-E) depending on diverse bacterial and archeal repeat sequences, as genes, and their mode of action ( 54 ). Type I and III systems share a common mechanism of processing of pre-crRNAs (to crRNA) via specialized Cas endonucleases, and on maturation, each crRNA complex with multi-Cas protein is capable of recognizing and cleaving target sequences which are complementary to crRNA. In contrary to this, Type II system is considered the heart of genome engineering tool because it involves reduced number of Cas enzymes ( Fig. 1 ).

Natural mechanisms of microbial adaptive immune CRISPR Systems: 1. Phage infection, 2. Spacer acquisition, 3. cRNA biogenesis, and processing ( 52 )
It requires non-coding tracrRNA which triggers the processing of pre-crRNA via dsRNA specific Ribonuclease RNase III and cas9 protein (only protein carrying out effective crRNA-mediated silencing of target) ( 55 ). CRISPR loci are predominantly composed of multiple repeated sequences (approx. 21–48 bp) interspaced by variable spacer sequences (approx. of 2–72 bp) and Cas genes situated alongside CRISPR locus. First, this CRISPR locus array is transcribed as single RNA, processing of this released pre-RNA (from within repeat sequences) into singular shorter units of CRISPR RNAs (crRNAs) using host proteins is done. Mature crRNA effectively binds to nucleases i.e. Cas proteins, thus this complex of crRNA-cas9 helps in recognizing and then cleaving the target invading DNA or RNA having complementarity to crRNA (sites called protospacers). A motif of 2–5 nucleotide called as PAM is located alongside protospacer in CRISPR-Cas systems I & II ( 54 ). PAM, a nucleic acid sequence made up of NGG or NAG trinucleotide for Cas9, flanks at 3’ end of the DNA target site, helps Cas9 in its specific cleavage activity and also facilitates Cas9 in distinguishing self-versus non self-bacterial sequences as PAM ( 52 ). SpCas9 (called as Streptococcus pyogenes Cas9) is being in use currently for effective genome editing in eukaryotic organisms including humans. PAM located downstream of target is only sequence allowing cas9 target site selection. Cas9 proteins vary in their size and sequence and have common domains as HNH and RuvC endonuclease to cleave two strands of target DNA ( Fig. 2 ).

Genome editing via CRISPR-Cas9 system. a) Double-strand breaks in DNA sequence can be repaired by cellular DNA repair mechanisms: the non-homologous end joining (NHEJ) or homology-directed repair (HDR)
Cas9HNH domain is specified for cleavage of strand complementary to guide (target) sequence while Cas9 RuvC domains for non-complementary (non-target) strand, In addition, Cas9 holds some conserved arginine-rich sites for binding of nucleic acid ( 53 ). Target recognition by this crRNA directs the silencing of foreign sequences by means of case proteins that function in complex with crRNAs ( 52 , 54 ).
The Streptococcus pyogenes-derived CRISPR–Cas9 RNA-guided DNA endonuclease is localized to a specific DNA sequence via a single guide RNA (sgRNA) sequence, which base pairs with a specific target sequence that is adjacent to a protos-pacer adjacent motif (PAM) sequence in the form of NGG or NAG.
On induction of double-stranded breaks or nicks at targeted regions, repairing is done by either Non-homologous end joining (NHEJ) or Homology-directed repair (HDR) pathway . NHEJ is an error-prone repair mechanism where joining of broken ends takes place, which generally results in heterogeneous indels (insertions and deletions) whereas HDR is a precise repair method in which homologous donor template DNA is being used in repair DNA damage target site ( 53 ).
Advantages of CRISPR over traditional methods
The CRISPR/Cas9 system is preferred over the ZNFs and TALENs because of many advantages. Firstly, the target design process is simpler for CRISPR as it depends upon the ribonucleotide complex formation instead of DNA recognition. It can be designed easily and it is much cheaper than designing nucleases as this does not need different proteins for each target and eliminates laborious cloning steps. This can be used to target any specific sequence in the genome. The CRISPR system is much more efficient than ZFNs and TALENs. The RNA encoding Cas protein can be injected directly for modifying the host genome. It is not so lengthy and laborious process as compared to traditional methods ( 12 , 51 ). By using CRISPR, we can introduce multiplexed mutations. Many genes can be mutated simultaneously by injecting with many gRNAs. This process is faster as compared to other methods. It does not introduce sensitivity to DNA methylation so it can be used if the target site is GC rich.
Molecular biology has rapidly evolved in the last decade than it has ever before. Different cancer treatment techniques are emerging and succeeding and with the development of ZNFs, TALENs, and CRISPR, scientists are able to target any sequence in the genome, even multiple genes. This will provide immense help in the treatment of diseases like cancer, avoiding the risks caused by the previous methods.
Ethical considerations
Ethical issues (Including plagiarism, informed consent, misconduct, data fabrication and/or falsification, double publication and/or submission, redundancy, etc.) have been completely observed by the authors.
Acknowledgments
The study was carried without financial support from any government or other educational organization. The authors acknowledged Dr. Idrees Ahmad Nasir and Dr. Muhammad Farooq Sabar for helping in editing of manuscript.
Conflict of interests
The authors declare that there is no conflict of interest.
share this!
April 23, 2024
This article has been reviewed according to Science X's editorial process and policies . Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
A universal framework for spatial biology
by Luca Marconato, Kevin Domanegg and Lisa Vollmar, European Molecular Biology Laboratory
Biological processes are framed by the context they take place in. A new tool developed by the Stegle Group from EMBL Heidelberg and the German Cancer Research Center (DKFZ) helps put molecular biology research findings in a better context of cellular surroundings, by integrating different forms of spatial data.
In a tissue, every individual cell is surrounded by other cells, and they all constantly interact with each other to give rise to biological function. To understand how tissues work or malfunction in diseases such as cancer, it is crucial to not only learn the characteristics of every cell, but also account for their spatial context. Quantitative characterization of cells in the context of the physical space they inhabit is key to understanding complex systems.
The technologies enabling these types of exploration are called spatial omics technologies, and their progressing development is contributing to the rise in popularity of spatial biology. Such technologies can give detailed information about the molecular makeup of individual cells and their spatial arrangement.
However, these technologies focus on different characteristics of a cell—such as RNA or protein levels, and the resulting datasets are managed and stored in diverse ways. To solve this challenge, a collaborative project led by the Stegle Group developed SpatialData, a data standard and software framework which allows scientists to represent data from a wide range of spatial omics technologies in a unified manner.
Technology development for spatial biology
Over the last decade, numerous technologies have been developed by both academia and industry for spatially visualizing tissues, cells, and subcellular compartments. However, each technique focuses on a small number of desirable characteristics and presents related trade-offs. For instance, Visium from 10x Genomics captures information about the expression of all genes in a tissue, but does not provide single-cell resolution.
In contrast, the 10x Genomics Xenium assay, MERFISH, or the MERSCOPE platform from Vizgen yield fine-grained maps of gene expression with subcellular resolution. However, these assays are currently limited to a few hundred preselected genes. And the list of such technologies, each providing a small slice of the full picture, keeps growing.
Challenges of spatial omics technologies
This heterogeneity of technologies is reflected on the computational side by an even greater heterogeneity of file formats: each technology comes with its own storage format, and often data generated by the same technology can be stored in multiple formats.
Practically, this brings several challenges to the analysis of spatial omics data. Visualization and analysis methods are usually tailored to a specific technology, which limits data compatibility and makes it hard to integrate different methods into a single analysis pipeline. However, for a holistic understanding of a biological system, it's important to simultaneously look at different cell characteristics or samples from different locations.
Omics technologies generate enormous amounts of data (terabytes of images, millions of cells, billions of single molecules), demanding optimized engineering solutions. Hence, spatial biology urgently needs a universal framework that can integrate data across experiments and technologies, and provide holistic insights into health and disease. This is where SpatialData steps in.
SpatialData—a framework to unite them all
"There is a strong need to establish community solutions for the management and storage of spatial omics data. In particular, there is a need to develop new data standards and computational foundations that allow for unifying analysis approaches across the full spectrum of different spatial omics technologies that are emerging," said Oliver Stegle, Group Leader at EMBL in the Genome Biology Unit, and head of the Computational Genomics and Systems Genetics division at the German Cancer Research Center (DKFZ).
"A first major step in this direction is SpatialData, a data standard and software framework that bridges and adapts previous data management concepts from single-cell multi-omics to the spatial domain."
SpatialData unifies and integrates data from different omics technologies, bridging state-of the-art-technologies with a framework that allows for computationally performant access and manipulation of the data.
This tool was introduced in a Nature Methods publication , authored by Luca Marconato during his Ph.D. at EMBL in the Stegle Group, a joint degree with the Faculty of Bioscience of the University of Heidelberg.
"We developed the SpatialData framework to alleviate the data representation challenges when studying spatial biology, so that the researcher can focus on the biological analysis, rather than being slowed down by tedious data manipulations, otherwise required to even just visualize the data. The framework provides a unified representation and implements ergonomic operations for convenient processing of spatial omics data," said Marconato.
The tool enables any researcher to import their data and perform tasks like data representation, processing, and visualization. Additionally, it gives the option to interactively annotate the data, and save it in a language-agnostic format, facilitating the emergence of analysis strategies that combine methods from different programming languages or analysis communities.
The framework has been developed as a collaborative project between multiple institutions such as the DKFZ, the Technical University of Munich, the Helmholtz Center Munich, German BioImaging, the ETH Zürich, VIB Center for Inflammation Research in Belgium, as well as the Huber and Saka groups at EMBL.
"We have conducted our research and technological development keeping the benefit for the bigger science community in mind," said Giovanni Palla, co-first author and Ph.D. student at the Helmholtz Center Munich.
"We not only established an interdisciplinary collaboration project between research institutes but also worked closely with developers working with different spatial technologies and in different programming languages to address the problem of interoperability. As a result, our framework is compatible with the vast majority of spatial omics assays from academia and industry.
"Being published openly, other researchers can now freely use SpatialData to manage their own data and have the opportunity to collaborate across various technologies and research topics."
"In our paper, we illustrate three important features of SpatialData," explained Kevin Yamauchi, co-first author and a postdoctoral researcher at ETH Zürich.
"First, we present a standardized interface and unified storage format (based on the OME-NGFF) for all spatial omics technologies. Second, using the unified representation, we integrate signals from multiple modalities. Here, we transfer annotations across modalities and quantify signals using these transferred annotations. Finally, we present a way to interactively annotate (pathology) images and use the annotations to analyze the associated molecular profiles."
SpatialData provides an interactive representation of data, both on your hard drive and your computer's RAM, which enables the analysis of large imaging data or multiple geometries or cells.
Other prominent key features are the framework's ability to align and annotate omics data in a common coordinate system. Thus, SpatialData enables the efficient management and manipulation of spatial datasets, including the definition of a common coordinate system across sequencing- and imaging-based technologies.
Application in breast cancer
The interdisciplinary team used the SpatialData framework to reanalyze a multimodal breast cancer dataset from 10X Genomics as a proof of concept. This dataset comprises consecutive sections of the same breast cancer block, where each section is analyzed using different technology, like Visium, Xenium, and a separate scRNA-seq dataset.
The study demonstrates the complementary nature of these technologies. "By integrating 10X Xenium and scRNAseq, we mapped the cell types into the space," said Elyas Heidari, a Ph.D. candidate at DKFZ and one of the authors of the study.
"Next, we used 10X Visium to identify cancer clones in space. This can be done because we have transcriptome-wide readouts. Finally, we used the H&E stained microscopy images to identify regions of interest for histopathology annotations. This analysis successfully showcased a unique application of SpatialData in unlocking multi-modal analyses of spatially-resolved datasets."
In the future, a patient's tumor might be analyzed with different technologies commonly used in the clinic, with the data then unified by SpatialData to gain a holistic understanding of the tumor. Furthermore, the interactive interface would allow the doctor to annotate the data, thus enabling detailed analysis of specific tumor regions and characteristics, potentially leading to personalized treatment approaches.
Journal information: Nature Methods
Provided by European Molecular Biology Laboratory
Explore further
Feedback to editors

Researchers find oldest undisputed evidence of Earth's magnetic field
39 minutes ago
Modeling broader effects of wildfires in Siberia

Scientists at the MAJORANA Collaboration look for rule-violating electrons

Lunar landforms indicate geologically recent seismic activity on the moon
2 hours ago

Japan's moon lander wasn't built to survive a weekslong lunar night. It's still going after 3
5 hours ago

Bioluminescence first evolved in animals at least 540 million years ago, pushing back previous oldest dated example
13 hours ago

Star bars show universe's early galaxies evolved much faster than previously thought
14 hours ago

Scientists study lipids cell by cell, making new cancer research possible

Squids' birthday influences mating: Male spear squids shown to become 'sneakers' or 'consorts' depending on birth date

Study finds rekindling old friendships as scary as making new ones
16 hours ago
Relevant PhysicsForums posts
The cass report (uk).
15 hours ago
Major Evolution in Action
Apr 22, 2024
If theres a 15% probability each month of getting a woman pregnant...
Apr 19, 2024
Can four legged animals drink from beneath their feet?
Apr 15, 2024
Mold in Plastic Water Bottles? What does it eat?
Apr 14, 2024
Dolphins don't breathe through their esophagus
More from Biology and Medical
Related Stories
Simplifying complexities in bioinformatics: A desktop suite for multi-omics data analysis and visualization
Mar 7, 2024
Benchmarking tool capable of closely mimicking single-cell and spatial genomics data
May 11, 2023

LIBRA: An adaptative integrative tool for paired single-cell multi-omics data
Nov 28, 2023

A new deep-learning-based analysis toolkit for spatial transcriptomics
Jan 2, 2024

Predicting cell fates: Researchers develop AI solutions for next-gen biomedical research
Feb 2, 2022

Chemical and computational advances enable 2D proteomics measurements of spatial signals
Feb 5, 2024
Recommended for you

World's chocolate supply threatened by devastating virus
19 hours ago

New small molecule helps scientists study regeneration
21 hours ago

A new method for enzymatic synthesis of potential RNA therapeutics

Researchers report on mechanisms of gene regulatory divergence between species

Bacteria for climate-neutral chemicals of the future
Researchers uncover natural variation in wild emmer wheat for broad-spectrum disease resistance
22 hours ago
Let us know if there is a problem with our content
Use this form if you have come across a typo, inaccuracy or would like to send an edit request for the content on this page. For general inquiries, please use our contact form . For general feedback, use the public comments section below (please adhere to guidelines ).
Please select the most appropriate category to facilitate processing of your request
Thank you for taking time to provide your feedback to the editors.
Your feedback is important to us. However, we do not guarantee individual replies due to the high volume of messages.
E-mail the story
Your email address is used only to let the recipient know who sent the email. Neither your address nor the recipient's address will be used for any other purpose. The information you enter will appear in your e-mail message and is not retained by Phys.org in any form.
Newsletter sign up
Get weekly and/or daily updates delivered to your inbox. You can unsubscribe at any time and we'll never share your details to third parties.
More information Privacy policy
Donate and enjoy an ad-free experience
We keep our content available to everyone. Consider supporting Science X's mission by getting a premium account.
E-mail newsletter
Maternal factor Trim75 contributes to zygotic genome activation program in mouse early embryos
- Original Article
- Published: 20 April 2024
- Volume 51 , article number 560 , ( 2024 )
Cite this article

- Weibo Hou 1 na1 ,
- Lijun Chen 1 na1 ,
- Jingzhang Ji 1 na1 ,
- Songling Xiao 1 ,
- Hongye Linghu 1 ,
- Lixin Zhang 1 ,
- Yue Ping 3 ,
- Chunsheng Wang 3 ,
- Qingran Kong 1 ,
- Wenpin Cai 2 &
- Xu Yang ORCID: orcid.org/0009-0005-1359-7613 1
17 Accesses
Explore all metrics
Zygotic genome activation (ZGA) is an important event in the early embryo development, and human embryo developmental arrest has been highly correlated with ZGA failure in clinical studies. Although a few studies have linked maternal factors to mammalian ZGA, more studies are needed to fully elucidate the maternal factors that are involved in ZGA.
Methods and results
In this study, we utilized published single-cell RNA sequencing data from a Dux-mediated mouse embryonic stem cell to induce a 2-cell-like transition state and selected potential drivers for the transition according to an RNA velocity analysis.
Conclusions
An overlap of potential candidate markers of 2-cell-like-cells identified in this research with markers generated by various data sets suggests that Trim75 is a potential driver of minor ZGA and may recruit EP300 and establish H3K27ac in the gene body of minor ZGA genes, thereby contributing to mammalian preimplantation embryo development.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Data availability
All sequencing data of mouse embryos generated in this study have been deposited in GEO under accession GSE227385.
Anifandis G, Messinis CI, Dafopoulos K, Messinis IE (2015) Genes and conditions Controlling mammalian pre- and post-implantation embryo development. Curr Genom 16(1):32–46
Article CAS Google Scholar
Jukam D, Shariati SAM, Skotheim JM (2017) Zygotic Genome Activation Vertebrates Dev Cell 42(4):316–332
CAS PubMed Google Scholar
Ko MS (2016) Zygotic genome activation revisited: looking through the expression and function of Zscan4. Curr Top Dev Biol 120:103–124
Article CAS PubMed Google Scholar
Svoboda P (2018) Mammalian zygotic genome activation. Semin Cell Dev Biol 84:118–126
De Iaco A et al (2017) DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 49(6):941–945
Article PubMed PubMed Central Google Scholar
Chen Z, Xie Z, Zhang Y (2021) DPPA2 and DPPA4 are dispensable for mouse zygotic genome activation and pre-implantation development . Development, 148(24)
Hu Z et al (2020) Maternal factor NELFA drives a 2 C-like state in mouse embryonic stem cells. Nat Cell Biol 22(2):175–186
Evans MJ, Kaufman MH (1981) Establishment in culture of pluripotential cells from mouse embryos. Nature 292(5819):154–156
Eckersley-Maslin MA, Alda-Catalinas C, Reik W (2018) Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat Rev Mol Cell Biol 19(7):436–450
Shen H et al (2021) Mouse totipotent stem cells captured and maintained through spliceosomal repression. Cell 184(11):2843–2859e20
Hu Y et al (2022) Induction of mouse totipotent stem cells by a defined chemical cocktail . Nature
Svensson V, Pachter L (2018) RNA Velocity: Molecular Kinetics from Single-Cell RNA-Seq Mol Cell. 72(1):7–9
Fu X et al (2019) Myc and Dnmt1 impede the pluripotent to totipotent state transition in embryonic stem cells. Nat Cell Biol 21(7):835–844
Article CAS PubMed PubMed Central Google Scholar
Xiong Z et al (2022) Ultrasensitive Ribo-Seq reveals translational landscapes during mammalian oocyte-to-embryo transition and pre-implantation development. Nat Cell Biol 24(6):968–980
Margalit L et al (2020) Trim24 and Trim33 play a role in epigenetic silencing of Retroviruses in embryonic stem cells . Viruses, 12(9)
Miao C et al (2021) TRIM37 orchestrates renal cell carcinoma progression via histone H2A ubiquitination-dependent manner. J Exp Clin Cancer Res 40(1):195
Li J et al (2022) Metabolic control of histone acetylation for precise and timely regulation of minor ZGA in early mammalian embryos. Cell Discov 8(1):96
Spanos KHaS (2002) Growth factor expression and function in the human and mouse preimplantation embryo. J Endocrinol 172:221–236
Article PubMed Google Scholar
Daniel A, Rappolee KSS, 0le Behrendtsen GA, Schultz RA, Pedersen, Werb Z (1992) Insulin-like growth factor II acts through an endogenous growth pathway regulated by imprinting m early mouse embryos, vol 6. GENES AND DEVELOPMENT, pp 939–952
Chegini TLPaN (1994) Immunohistochemical localization of insulin-like growth factor (IGF-I), IGF-I receptor, and IGF binding proteins 1–4 in human fallopian tube at various reproductive stages, vol 50. BIOLOGY OF REPRODUCTION, pp 281–289
Alandardik RMS, Schultz R (1992) Colocalization of transforming growth factor-alpha and a functional epidermal growth factor receptor (EGFR) to the inner cell mass and preferential localization of the EGFR on the basolateral surface of the trophectoderm in the mouse blastocyst DEVELOPMENTAL BIOLOGY. 154:396–409
Chia CM, Handyside RMWAH (1995) EGF, TGF-alpha and EGFR expression in human preimplantation embryos. DEVELOPMENT 121(2):299–307
Aoki F (2022) Zygotic gene activation in mice: profile and regulation. J Reprod Dev 68(2):79–84
Rodriguez-Terrones D et al (2020) A distinct metabolic state arises during the emergence of 2-cell-like cells. EMBO Rep 21(1):e48354
Sang Q et al (2021) Genetic factors as potential molecular markers of human oocyte and embryo quality. J Assist Reprod Genet 38(5):993–1002
Download references
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Weibo Hou, Lijun Chen and Jingzhang Ji contributed equally to this work and share first authorship.
Authors and Affiliations
School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, Zhejiang Province, China
Weibo Hou, Lijun Chen, Jingzhang Ji, Songling Xiao, Hongye Linghu, Lixin Zhang, Qingran Kong & Xu Yang
Department of Laboratory Medicine, Wenzhou Hospital of Traditional Chinese Medicine Affiliated to Zhejiang Chinese Medicine University, Wenzhou, Zhejiang, People’s Republic of China
College of Life Science, Northeast Forestry University, No. 26, hexing Road, Harbin, China
Yue Ping & Chunsheng Wang
You can also search for this author in PubMed Google Scholar
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Weibo Hou, Lijun Chen, Jingzhang Ji, Songling Xiao, Hongye Linghu, Lixin Zhang, Yue Ping and Chunsheng Wang. The first draft of the manuscript was written by Xu Yang and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Correspondence to Qingran Kong , Wenpin Cai or Xu Yang .
Ethics declarations
Ethical approval.
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher’s note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 1
Supplementary material 2, rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Hou, W., Chen, L., Ji, J. et al. Maternal factor Trim75 contributes to zygotic genome activation program in mouse early embryos. Mol Biol Rep 51 , 560 (2024). https://doi.org/10.1007/s11033-024-09349-0
Download citation
Received : 28 June 2023
Accepted : 12 February 2024
Published : 20 April 2024
DOI : https://doi.org/10.1007/s11033-024-09349-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Preimplantation embryo development
- Find a journal
- Publish with us
- Track your research
HYPOTHESIS AND THEORY article
This article is part of the research topic.
Using Case Study and Narrative Pedagogy to Guide Students Through the Process of Science
Molecular Storytelling: A Conceptual Framework for Teaching and Learning with Molecular Case Studies Provisionally Accepted

- 1 School of Interdisciplinary Arts and Sciences, University of Washington Bothell, United States
- 2 Institute for Quantitative Biomedicine, Rutgers, The State University of New Jersey, United States
- 3 Research Collaboratory for Structural Bioinformatics Protein Data Bank, Rutgers, The State University of New Jersey,, United States
The final, formatted version of the article will be published soon.
Molecular case studies (MCSs) provide educational opportunities to explore biomolecular structure and function using data from public bioinformatics resources. The conceptual basis for the design of MCSs has yet to be fully discussed in the literature, so we present molecular storytelling as a conceptual framework for teaching with case studies. Whether the case study aims to understand the biology of a specific disease and design its treatments or track the evolution of a biosynthetic pathway, vast amounts of structural and functional data, freely available in public bioinformatics resources, can facilitate rich explorations in atomic detail. To help biology and chemistry educators use these resources for instruction, a community of scholars collaborated to create the Molecular CaseNet. This community uses storytelling to explore biomolecular structure and function while teaching biology and chemistry. In this article, we define the structure of an MCS and present an example. Then, we articulate the evolution of a conceptual framework for developing and using MCSs. Finally, we related our framework to the development of technological, pedagogical, and content knowledge (TPCK) for educators in the Molecular CaseNet. The report conceptualizes an interdisciplinary framework for teaching about the molecular world and informs lesson design and education research.
Keywords: Molecular education, Case studies, Technological pedagogical and content knowledge (TPCK), Molecular structure and function, molecular visualization, Bioinformatics education, conceptual modeling
Received: 31 Jan 2024; Accepted: 23 Apr 2024.
Copyright: © 2024 Trujillo and Dutta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY) . The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
* Correspondence: Prof. Caleb M. Trujillo, University of Washington Bothell, School of Interdisciplinary Arts and Sciences, Bothell, United States
People also looked at
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
Molecular biology articles within Nature Reviews Genetics
Journal Club | 24 April 2024
What tubulin can teach us about gene regulation
In this Journal Club article, Olivia Rissland describes how a 1987 paper by Don Cleveland and colleagues provided insight into co-translational gene regulation of tubulin.
- Olivia S. Rissland
Review Article | 18 April 2024
Nuclear mRNA decay: regulatory networks that control gene expression
In this Review, the authors summarize our current understanding of nuclear pre-mRNA and mRNA decay pathways. They describe how aberrantly processed mRNAs are targeted for decay in the nucleus and how this process is regulated to finely control gene expression.
- Xavier Rambout
- & Lynne E. Maquat
Perspective | 17 April 2024
Natural antisense transcripts as versatile regulators of gene expression
In this Perspective, Werner and colleagues discuss the many potential mechanisms by which natural antisense transcripts (NATs) can regulate expression of their complementary sense transcripts, the biological implications of their regulatory effects and the potential of NATs for therapeutic applications.
- Andreas Werner
- , Aditi Kanhere
- & John S. Mattick
Research Highlight | 12 April 2024
A developmental exit from totipotency
A paper in Nature Genetics identifies a mechanism involving the transcription factor DUXBL that controls the development of early embryonic mouse cells past stages marked by totipotency.
Tools of the Trade | 12 April 2024
SIMPLE-seq to decode DNA methylation dynamics in single cells
In this Tools of the Trade article, Dongsheng Bai and Chenxu Zhu describe SIMPLE-seq, a scalable single-cell sequencing method that simultaneously decodes the cytosine modifications 5mC and 5hmC.
- Dongsheng Bai
- & Chenxu Zhu
Research Highlight | 12 March 2024
Competition between sites of meiotic recombination in snakes
A study in Science reports that corn snakes use both PRDM9 and promoter-like features to direct meiotic recombination, indicating that these are not mutually exclusive.
- Kirsty Minton
Journal Club | 27 February 2024
The continuum of transcription factor affinities
Carl G. de Boer highlights a recent paper by Lim et al. on the importance low-affinity transcription factor-binding sites for determining organismal phenotypes.
- Carl G. de Boer
Journal Club | 30 January 2024
Transcriptional diversity along the intestinal crypt–villi axis
Kylie R. James discusses a paper by Moor et al. that described a transcriptional gradient along the intestinal axis using single-cell RNA sequencing.
- Kylie R. James
Review Article | 09 January 2024
Real-time single-molecule imaging of transcriptional regulatory networks in living cells
In this Review, Hwang and co-authors outline single-molecule fluorescence imaging techniques that can be used in living cells to visualize individual molecules involved in the spatiotemporal regulation of gene expression. This Review also delves into the biological insights gained through these methodologies.
- Dong-Woo Hwang
- , Anna Maekiniemi
- & Hanae Sato
Review Article | 30 November 2023
Divergence and conservation of the meiotic recombination machinery
In this Review, the authors describe the evolutionary conservation and divergence of the meiotic recombination machinery, focusing on proteins that are required for meiotic double-strand break formation, double-strand break repair via homologous recombination and the formation of crossover and non-crossover recombinant DNA molecules.
- Meret Arter
- & Scott Keeney
Research Highlight | 27 November 2023
SWI/SNF function compensated by another chromatin remodeller
Martin et al. report that the EP400/TIP60 complex can compensate for function of the SWI/SNF complex to remodel chromatin at some promoters.
Review Article | 24 November 2023
Context-specific functions of chromatin remodellers in development and disease
In this Review, the authors summarize the biological roles of chromatin remodellers and describe the complex mechanisms that underpin their specific functions, with an emphasis on evidence from large-scale genetic studies.
- Sai Gourisankar
- , Andrey Krokhotin
- & Gerald R. Crabtree
Review Article | 15 November 2023
Non-coding RNAs in disease: from mechanisms to therapeutics
In this Review, the authors describe our current knowledge of the role of microRNAs, long non-coding RNAs and circular RNAs in disease, with a focus on cardiovascular, neurological, infectious diseases and cancer. Further, they discuss the potential use of non-coding RNAs as disease biomarkers and as therapeutic targets.
- Kinga Nemeth
- , Recep Bayraktar
- & George A. Calin
Viewpoint | 22 September 2023
Pioneer factors — key regulators of chromatin and gene expression
Five leading researchers provide their perspectives on our current understanding of pioneer factors and their important gene regulatory roles in cell differentiation, cell fate determination and reprogramming.
- Martha L. Bulyk
- , Jacques Drouin
- & Kenneth S. Zaret
Review Article | 06 September 2023
Computational methods for analysing multiscale 3D genome organization
In this Review, Zhang et al. discuss how recent advances in computational methods are helping to reveal the multiscale features involved in genome folding within the nucleus and how the resulting 3D genome organization relates to genome function.
- , Lorenzo Boninsegna
- & Jian Ma
Review Article | 02 August 2023
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
In this Review, the authors discuss recent advances in our understanding of Mediator and TFIID, coactivators associated with the RNA polymerase II (Pol II) pre-initiation complex (PIC), focusing on their structure, interactions with activators and impact on the function of the PIC.
- Sohail Malik
- & Robert G. Roeder
Research Highlight | 28 July 2023
Spatial mapping of translation in single cells
Zeng et al. introduce RIBOmap, a three-dimensional in situ imaging technique that enables the mapping of mRNAs undergoing translation.
- Michael Attwaters
Journal Club | 25 July 2023
The chromosome hoarding syndrome of (some) ferns and lycophytes
Fay-Wei Li recalls a 1966 paper by Klekowski and Baker, who built on their observation that homosporous pteridophytes have many more chromosomes than heterosporous lineages to generate hypotheses on the evolutionary impact of polyploidy.
Review Article | 28 June 2023
microRNAs in action: biogenesis, function and regulation
In this Review, the authors describe how the application of new technologies to the microRNA (miRNA) field has yielded key insights into miRNA biology. The authors summarize our current understanding of miRNA biogenesis, function and processing, and highlight challenges to address in future research.
- Renfu Shang
- , Seungjae Lee
- & Eric C. Lai
Review Article | 07 June 2023
Transposable elements in mammalian chromatin organization
This Review discusses how transposable elements contribute to mammalian genome evolution and gene regulation through their ability to both maintain and reshape 3D genome structure.
- Heather A. Lawson
- , Yonghao Liang
- & Ting Wang
Research Highlight | 17 May 2023
Chromatin loops stack up
A study in Molecular Cell reveals how CTCF and cohesin contribute to genome organization through their roles in forming and stacking chromatin loops.
- Dorothy Clyde
Review Article | 12 May 2023
Deafness: from genetic architecture to gene therapy
The authors review genetic studies of sensorineural hearing impairment (SNHI) and their resulting insights into the molecular mechanisms underlying auditory system function. They also discuss preclinical studies of inner-ear gene therapy and key translational opportunities and challenges for treating monogenic forms of SNHI and associated balance disorders.
- Christine Petit
- , Crystel Bonnet
- & Saaïd Safieddine
Review Article | 11 May 2023
The epithelial–mesenchymal plasticity landscape: principles of design and mechanisms of regulation
Recent systems biology and single-cell approaches have revealed the impact of the microenvironment, lineage specification and cell identity, and the genome on epithelial–mesenchymal plasticity (EMP). In addition, cell memory (hysteresis) and cellular noise can drive stochastic transitions between cell states. The authors review these forces and the regulatory mechanisms that stabilize EMP states or facilitate epithelial–mesenchymal transitions (EMTs).
- Jef Haerinck
- , Steven Goossens
- & Geert Berx
Research Highlight | 10 May 2023
microRNAs as systemic regulators of ageing
Wagner et al. report an organism-wide map of non-coding RNA expression in ageing and rejuvenated mice, identifying a set of broadly deregulated microRNAs that may act as systemic regulators of ageing.
Review Article | 09 May 2023
Interpreting non-coding disease-associated human variants using single-cell epigenomics
In this Review, Gaulton et al. discuss how single-cell epigenomic methods generate cell type-, subtype- and state-resolved maps of candidate cis -regulatory elements in heterogeneous human tissues that can help to interpret the genetic basis of common traits and diseases.
- Kyle J. Gaulton
- , Sebastian Preissl
- & Bing Ren
Research Highlight | 05 May 2023
The different faces of transcription factor sensitivity
A new study in Nature Genetics describes how changes in the levels of transcription factors can affect normal-range phenotypic variation and disease.
Research Highlight | 28 April 2023
Cryptic initiation drives transcriptional junk in ageing
Sen et al. describe the epigenetic changes that underpin aberrant transcription initiation during senescence and ageing.
Review Article | 02 March 2023
Methods and applications for single-cell and spatial multi-omics
In this Review, the authors discuss the latest advances in profiling multiple molecular modalities from single cells, including genomic, transcriptomic, epigenomic and proteomic information. They describe the diverse strategies for separately analysing different modalities, how the data can be computationally integrated, and approaches for obtaining spatially resolved data.
- Katy Vandereyken
- , Alejandro Sifrim
- & Thierry Voet
Research Highlight | 28 February 2023
Engineering transgenerational epigenetic inheritance in mammals
A new study in Cell uses epigenome engineering to confer transgenerational inheritance of DNA methylation states and metabolic traits in mice.
- Darren J. Burgess
Journal Club | 06 February 2023
Regulatory promoter architectures in the hands of thermodynamic modelling
Julio Collado-Vides recalls two 2005 publications that provide a conceptual framework based on a statistical thermodynamics approach to quantitatively model the regulatory activity at promoters subject to regulation by multiple transcription factors.
- Julio Collado-Vides
Journal Club | 02 February 2023
Chromatin organization in red triangles
This journal club by Elisa Oricchio highlights two studies published in 2012, which used chromatin conformation capture methods to detect the formation of self-interacting chromatin regions, known as topologically associating domains (TADs).
- Elisa Oricchio
How Hi-C ignited the era of 3D genome biology
Magda Bienko highlights a landmark paper by Lieberman-Aiden et al., which in 2009 reported the development of high-throughput chromosome conformation capture (Hi-C), revolutionizing the field of 3D genome biology.
- Magda Bienko
Research Highlight | 30 January 2023
Exon junction complex modulates m 6 A distribution
Two studies have revealed that the characteristic distribution of N 6-methyladenosine (m 6 A) — an RNA modification known to be functionally important for mRNA metabolism among other processes — in mRNA is shaped by the exon junction complex during splicing.
Review Article | 30 January 2023
Molecular mechanisms of environmental exposures and human disease
Environmental pollutants have been shown to disrupt molecular mechanisms underlying common complex diseases. The authors review the interplay of environmental stressors with the human genome and epigenome as well as other molecular processes, such as production of extracellular vesicles, epitranscriptomic changes and mitochondrial changes, through which the environment can exert its effects.
- , Christina M. Eckhardt
- & Andrea A. Baccarelli
Review Article | 18 January 2023
Navigating the pitfalls of mapping DNA and RNA modifications
The ability to map DNA and RNA modifications has improved our understanding of these marks, but in some cases inconsistent results have been problematic. Here, Kong et al. discuss how to recognize and resolve issues associated with commonly used sequencing-based approaches to minimize mapping errors.
- Yimeng Kong
- , Edward A. Mead
- & Gang Fang
Review Article | 12 January 2023
Autophagy genes in biology and disease
Macroautophagy and microautophagy involve characteristic membrane dynamics regulated by autophagy-related proteins to degrade cytoplasmic material in lysosomes. In this Review, the authors summarize recent progress in elucidating these highly conserved processes, the pathological relevance of autophagy-related genes in Mendelian and complex diseases, and the evolution of the autophagy pathway.
- Hayashi Yamamoto
- , Sidi Zhang
- & Noboru Mizushima
Review Article | 16 December 2022
Regulation of pre-mRNA splicing: roles in physiology and disease, and therapeutic prospects
Alternative splicing of pre-mRNAs is key for cellular function and underpins the aetiology of numerous diseases. Here, we review major advances in understanding the structures and functions of the splicing machinery and its regulation, and in harnessing this knowledge for the design of novel therapies.
- Malgorzata Ewa Rogalska
- , Claudia Vivori
- & Juan Valcárcel
Review Article | 23 November 2022
The nexus between RNA-binding proteins and their effectors
This Review surveys the known mechanisms of communication between RBPs and their effectors and their roles in reducing the complexity of RNA networks. The authors review the emerging roles of RBP–effector interactions in the control of RNA processing and regulation of biological outcomes, and their contribution to human health and disease.
- , Eugene Valkov
- & Jernej Murn
Research Highlight | 14 November 2022
Locking in a synthetic genetic code
A new study in Science reports the refactoring of genetic codes in Escherichia coli to create a bidirectional ‘genetic firewall’ that prevents genetic transfer from or to synthetic organisms.
Review Article | 08 November 2022
Probing the dynamic RNA structurome and its functions
In this Review, Spitale and Incarnato discuss how the application of sequencing-based RNA structure mapping methods to entire transcriptomes in living cells is providing insight into the RNA structurome, the dynamics of RNA ensembles and how RNA structure regulates cellular processes.
- Robert C. Spitale
- & Danny Incarnato
Review Article | 31 October 2022
Dynamic alternative DNA structures in biology and disease
Non-B DNA secondary structures, such as G quadruplexes, H-DNA or Z-DNA, have key roles in genetic instability and disease aetiology. The authors review the impact of non-B DNA on transcription, replication, recombination and DNA damage and repair, the mechanisms of non-B DNA-induced mutagenesis and the role of non-B DNA sequences in human disease.
- Guliang Wang
- & Karen M. Vasquez
Research Highlight | 25 October 2022
RADARs and READRs for programmable RNA sensing
Two new papers report programmable systems that leverage ADAR adenosine deaminases for recording transcript abundance.
Review Article | 19 October 2022
Biological roles of adenine methylation in RNA
Boulias and Greer review the functions of N 6 -methyladenosine (m 6 A) in RNA at the molecular, genomic and organismal level. They describe the impact of m 6 A deposition on RNA substrates, chromatin architecture, epigenetic regulation of gene expression and genome stability, as well as key roles of m 6 A in stem-cell differentiation, neurogenesis and immunity.
- Konstantinos Boulias
- & Eric Lieberman Greer
In Brief | 07 October 2022
Target-directed microRNA degradation in Drosophila
A report in Molecular Cell identifies six new triggers of target-directed miRNA degradation, which are essential for normal Drosophila melanogaster embryogenesis.
Review Article | 30 September 2022
New insights into genome folding by loop extrusion from inducible degron technologies
Inducible protein degradation technologies enable the depletion of loop extrusion factors within short time frames, leading to the rapid reconfiguration of the 3D genome. Nora and de Wit review insights from degron approaches into the molecular factors controlling genome folding and how these findings have changed our understanding of genome organization, including its role in transcription.
- Elzo de Wit
- & Elphège P. Nora
Research Highlight | 27 September 2022
Promoting a new view of mitochondrial genome regulation
A paper in Molecular Cell reports the characterization of a second functional light strand promoter (LSP2) in the mitochondrial genome, challenging the view that mitochondrial DNA replication and gene expression are coupled by their reliance on a single light strand promoter (LSP).
Review Article | 14 September 2022
The spatial organization of transcriptional control
This Review discusses how chromosome tracing has deepened our understanding of the role of 3D chromatin topology in transcriptional regulation by helping to resolve open questions and opposing models rising from data generated by sequencing-based approaches, such as 3C and HiC.
- Antonina Hafner
- & Alistair Boettiger
Review Article | 20 July 2022
The emerging landscape of spatial profiling technologies
Spatial omics methods enable the charting of cellular heterogeneity, complex tissue architectures and dynamic changes during development and disease. The authors review the developing landscape of in situ spatial transcriptome, genome and proteome technologies and highlight their impact on basic and translational research.
- Jeffrey R. Moffitt
- , Emma Lundberg
- & Holger Heyn
Review Article | 12 July 2022
Generating specificity in genome regulation through transcription factor sensitivity to chromatin
In this Review, Isbel et al. describe our current understanding of how transcription factors navigate features of chromatin — particularly DNA methylation and nucleosomes — and how this contributes to specificity of genomic binding and, ultimately, transcriptional regulation.
- , Ralph S. Grand
- & Dirk Schübeler
Perspective | 12 July 2022
Alternative splicing as a source of phenotypic diversity
In this Perspective, the authors discuss how regulated alternative splicing can generate phenotypic diversity and outline emerging evidence that alternative splicing contributes to adaptation and species divergence.
- Charlotte J. Wright
- , Christopher W. J. Smith
- & Chris D. Jiggins
Browse broader subjects
- Biological sciences
Browse narrower subjects
- Cell division
- Chromosomes
- CRISPR-Cas systems
- DNA damage and repair
- DNA metabolism
- DNA recombination
- DNA replication
- Epigenetics
- Non-coding RNAs
- Nuclear organization
- Post-translational modifications
- Protein folding
- Proteolysis
- Riboswitches
- RNA metabolism
- Single-molecule biophysics
- Transcription
- Transcriptomics
- Translation
- Transposition
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies

COMMENTS
Molecular biology articles from across Nature Portfolio. Molecular Biology is the field of biology that studies the composition, structure and interactions of cellular molecules such as nucleic ...
Current research has shown that microRNAs (miRNAs) play vital roles in plant response to stress caused by heavy metals such as aluminum, arsenic, cadmium (Cd), and mercury. Cd has become one of the most hazard... Jian Gao, Mao Luo, Hua Peng, Fabo Chen and Wenbo Li. BMC Molecular Biology 2019 20 :14.
Molecular biology. Read the latest research on molecular biology or search thousands of news articles with images from leading universities and research institutes.
Genomic research has evolved from seeking to understand the fundamentals of the human genetic ... the Department of Molecular and Cell Biology and the Department of Chemistry, University of ...
Journal of Molecular Biology (JMB) provides high quality, comprehensive and broad coverage in all areas of molecular biology.The journal publishes original scientific research papers that provide mechanistic and functional insights and report a significant advance to the field. The journal encourages the submission of multidisciplinary studies that use complementary experimental and ...
Molecular biology articles within Nature. Featured. Article 10 April 2024 | Open Access. ... Research articles News Opinion Research Analysis Careers ...
Jonghui Kim, Karla Hegener, Claudia Hagedorn, Daniel Weidinger, Kashin Jamal Jameel, Inga Marte Charlott Seuthe, Sabine Eichhorn, Florian Kreppel, Jonas Jae-Hyun Park and Jürgen Knobloch. BMC Molecular and Cell Biology 2023 24 :31. Research Published on: 10 October 2023. Full Text.
A peer-reviewed monthly reviews journal that publishes cutting-edge authoritative and accessible reviews in the fields of molecular and cell biology.
Empowering a community publishing articles in Molecular Biology, including RNA biology, chromatin and epigenetics, gene expression, translation, DNA replication and repair, nuclear organization, gene editing, and much more. ... PLOS is a leader in Molecular Biology research. More than 32,735 articles. 979,496 citations. Authors from 151 countries.
Read the latest articles of Journal of Molecular Biology at ScienceDirect.com, Elsevier's leading platform of peer-reviewed scholarly literature ... Research article Open access CATH 2024: CATH-AlphaFlow Doubles the Number of Structures in CATH and Reveals Nearly 200 New Folds ... select article Molecular Insights into Aggrephagy: Their ...
PCR, Cloning, Restriction Digestion, Ligation, Transformation, Plasmid et al | Explore the latest full-text research PDFs, articles, conference papers, preprints and more on MOLECULAR BIOLOGY.
Abstract. In recent decades, advances in methods in molecular biology and genetics have revolutionized multiple areas of the life and health sciences. However, there remains a global need for the development of more refined and effective methods across these fields of research. In this current Collection, we aim to showcase articles presenting ...
Overview. Molecular Biology is a peer-reviewed journal covering various domains of molecular, cell, and computational biology. Offers wide-ranging coverage of problems related to molecular and cell biology. Encompasses genomics, proteomics, bioinformatics, molecular virology and immunology, molecular development biology, molecular evolution ...
The Protective Action of Hsp70 and Hydrogen Sulfide Donors in THP-1 Macrophages in the Lipopolysaccharide-Induced Inflammatory Response by Modulating Endocytosis. Molecular Biology is a peer-reviewed journal covering various domains of molecular, cell, and computational biology. Offers wide-ranging coverage of problems ...
BMC Molecular and Cell Biology, formerly known as BMC Cell Biology, is an open access journal that considers articles on all aspects of cellular and molecular biology in both eukaryotic and prokaryotic cells.The journal considers studies on functional cell biology, molecular mechanisms of transcription and translation, biochemistry, as well as research using both the experimental and ...
Read the latest articles of Journal of Molecular Biology at ScienceDirect.com, Elsevier's leading platform of peer-reviewed scholarly literature. Skip to main content. ADVERTISEMENT ... Research article Open access Membrane specificity of the human cholesterol transfer protein STARD4. Reza Talandashti, Larissa van Ek, Charlotte Gehin, Dandan ...
The 100 most downloaded cell and molecular biology research articles published in Scientific Reports in 2023 Skip to main content Thank you for visiting nature.com.
Molecular Biology Reports is a peer-reviewed general molecular biology journal publishing sound science research and Reviews in all areas of molecular and cellular biology.. Covers both eukaryote (animals, plants, algae, fungi) and prokaryote (bacteria and archaea) in vitro and in vivo research. Welcomes fundamental and translational research, as well as new techniques that advance ...
Integrating Evolutionary Paradigms into Cancer Research. Understanding cancer from the lens of evolutionary theory is essential to fully comprehend cancer's behavior. Herein we present a perspective on cancer and evolution that resulted from discussion during our SMBE-sponsored satellite meeting on the molecular biology and evolution of cancer.
Sarah Batey. Jane Clarke. Analysis 14 Mar 2007. Nature Reviews Molecular Cell Biology ( Nat Rev Mol Cell Biol) ISSN 1471-0080 (online) ISSN 1471-0072 (print) Read the latest Research articles from ...
This article was written from the data collected from well-reputed `databases of PubMed, Science Direct, Springer and Elsevier groups of journals. The data were collected in the form of research and review articles on the use of molecular biology for cancer research, which was summarized to form an improved and advance review article.
Read the latest Research articles from Nature Structural & Molecular Biology. ... Research articles. Filter By: Article Type. All. All; Analysis (17) Article (3327) Brief Communication (229)
Molecular Biology. I. Vitale, in Reference Module in Life Sciences, 2017 Impact of Molecular Biology in Life Science. The field of molecular biology has a profound impact in life science investigation. Major advances in molecular biology over the last four decades have stimulated research and progress in almost all the disciplines of life science.
A new tool developed by the Stegle Group from EMBL Heidelberg and the German Cancer Research Center (DKFZ) helps put molecular biology research findings in a better context of cellular ...
Molecular Biology Reports Aims and scope Submit manuscript ... An overlap of potential candidate markers of 2-cell-like-cells identified in this research with markers generated by various data sets suggests that Trim75 is a potential driver of minor ZGA and may recruit EP300 and establish H3K27ac in the gene body of minor ZGA genes, ...
To help biology and chemistry educators use these resources for instruction, a community of scholars collaborated to create the Molecular CaseNet. This community uses storytelling to explore biomolecular structure and function while teaching biology and chemistry. In this article, we define the structure of an MCS and present an example.
Recent systems biology and single-cell approaches have revealed the impact of the microenvironment, lineage specification and cell identity, and the genome on epithelial-mesenchymal plasticity ...