Academia Bees

How to Write Acknowledgement for Research Paper (5 Samples)
July 12, 2023
No Comments
By Mohsin Khurshid
Writing acknowledgements is an essential part of crafting a comprehensive research paper. It allows you to express gratitude and recognize the contributions of individuals and institutions who have supported your work. In this article, we will delve into the art of writing acknowledgement for research papers, providing you with valuable insights, practical tips, and five sample acknowledgements to guide you in acknowledging the people and resources that have played a significant role in your research journey.
Table of Contents
- 1 Understanding the Role of Acknowledgements in Research Papers
- 2 Key Elements of an Effective Acknowledgement
- 3 10 Tips for Writing an Acknowledgement for a Research Paper
- 4.1 Sample 1: Acknowledgement for Collaborative Research:
- 4.2 Sample 2: Acknowledgement for Funding Support:
- 4.3 Sample 3: Acknowledgement for Mentorship and Guidance:
- 4.4 Sample 4: Acknowledgement for Institutional Support:
- 4.5 Sample 5: Acknowledgement for Peer Reviewers:
- 6 Conclusion
Understanding the Role of Acknowledgements in Research Papers
Acknowledgements serve as a platform to express appreciation and recognize the collective effort that goes into the completion of a research paper. They provide an opportunity to acknowledge the guidance, support, and assistance received throughout the research process. By including acknowledgements, you can demonstrate your gratitude and give credit to those who have contributed to your success.
Key Elements of an Effective Acknowledgement
Crafting an effective acknowledgement involves considering various elements to ensure its sincerity and clarity. It is crucial to mention specific individuals, institutions, and their contributions, while keeping the acknowledgement concise and relevant. By adhering to ethical considerations and cultural norms, you can create an acknowledgement that reflects your gratitude and professionalism.
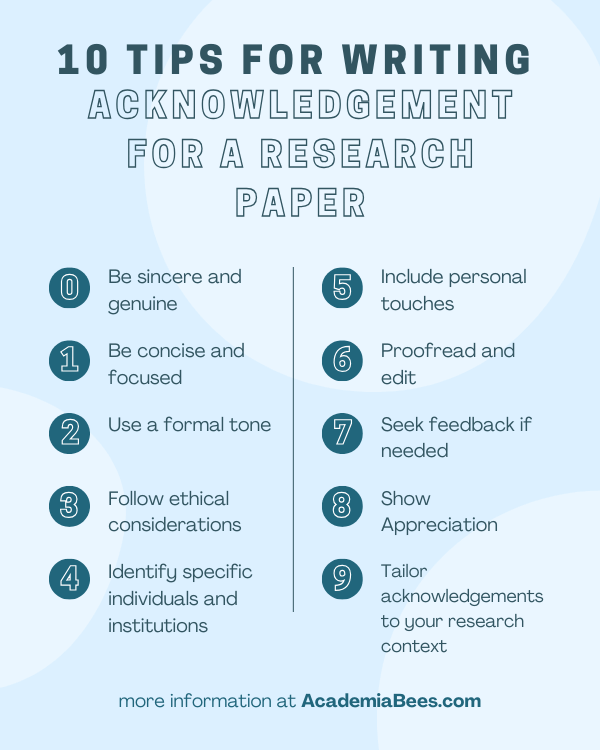
10 Tips for Writing an Acknowledgement for a Research Paper
- Be sincere and genuine : Write your acknowledgements with heartfelt gratitude, expressing sincere appreciation for the support and contributions received during your research.
- Identify specific individuals and institutions: Mention the names of people who have played a significant role in your research, such as mentors, advisors, collaborators, and funding agencies. Also, acknowledge the contributions of institutions that provided resources or facilities.
- Be concise and focused: Keep your acknowledgements concise and to the point. Focus on the key individuals and organizations that have made a substantial impact on your research.
- Use a formal tone: Maintain a professional and formal tone throughout your acknowledgements. Remember that this section is a formal acknowledgment of support, not a casual conversation.
- Follow ethical considerations: Ensure that you acknowledge individuals and organizations according to ethical guidelines and norms. Respect the privacy and confidentiality of individuals who may prefer not to be mentioned.
- Include personal touches: If appropriate, you can include personal anecdotes or specific instances where individuals or organizations made a significant impact on your research.
- Tailor acknowledgements to your research context: Consider the nature of your research and tailor your acknowledgements accordingly. For example, if you conducted interdisciplinary research, acknowledge experts from different fields who provided valuable insights.
- Proofread and edit: Like any other section of your research paper, proofread and edit your acknowledgements for grammar, spelling, and clarity. Ensure that the acknowledgements are well-written and free of errors.
- Seek feedback if needed: If you’re uncertain about whom to acknowledge or how to express your gratitude, seek feedback from your mentors, advisors, or colleagues. They can provide valuable guidance and suggestions.
- Show appreciation beyond formal requirements: While it’s important to acknowledge the required individuals and institutions, also consider extending your appreciation to others who may have supported you indirectly, such as family, friends, or colleagues who provided emotional support during your research journey.
Remember, acknowledgements are an opportunity to express your gratitude and recognize the contributions of those who have helped you along the way. Take the time to craft a thoughtful and sincere acknowledgement section that reflects the support and collaborative spirit of your research endeavor.
5 Samples for Acknowledgment in Research Paper
Explore these 5 carefully crafted acknowledgment samples to effectively express gratitude in your research paper.
Sample 1: Acknowledgement for Collaborative Research:
In this sample, we showcase an acknowledgement that acknowledges the collaborative efforts of research collaborators and team members. It highlights the importance of recognizing the joint contributions made towards the research project.
“I would like to express my deepest appreciation to the members of the research team, [Collaborators’ Names], for their invaluable contributions and collaborative spirit throughout this research project. Our collective efforts and synergistic teamwork have significantly enhanced the quality and depth of this study. Each member’s unique expertise and perspectives have brought forth diverse insights, resulting in a more comprehensive and well-rounded analysis.
I am grateful for the dedication, commitment, and professionalism demonstrated by each team member. The constructive discussions, intellectual debates, and shared enthusiasm have fostered an enriching research environment that has truly pushed the boundaries of our collective knowledge. This research project stands as a testament to the power of collaboration and the collective pursuit of knowledge.”
Sample 2: Acknowledgement for Funding Support:
This sample acknowledgement focuses on acknowledging the financial support received for the research. It emphasizes the significance of recognizing funding agencies or organizations that have provided the necessary resources for the research to take place.
“I would like to extend my sincere gratitude to the funding agencies and organizations that have provided financial support for this research. Their generous contributions have made it possible to conduct this study and have significantly contributed to its successful completion. The financial support has allowed for the procurement of necessary research materials, access to specialized equipment, and the opportunity to engage in valuable research experiences.
I would like to express my appreciation to [Name of Funding Agency/Organization 1] for their generous grant, which has played a crucial role in supporting this research project. Their belief in the significance of this study and their commitment to advancing knowledge in this field have been instrumental in its realization.
Furthermore, I would like to acknowledge the support received from [Name of Funding Agency/Organization 2]. Their funding has been vital in facilitating data collection, analysis, and the dissemination of research findings. Their investment in this project has not only provided financial resources but has also validated the importance and potential impact of this research.”
Sample 3: Acknowledgement for Mentorship and Guidance:
Here, we present a sample acknowledgement that expresses gratitude towards mentors and advisors who have provided guidance and support throughout the research journey. It underscores the critical role of mentorship in academic and research endeavors.
“I am deeply grateful to my mentor, [Mentor’s Name], for their exceptional guidance and unwavering support throughout this research endeavor. Their expertise, insightful feedback, and continuous encouragement have been invaluable in shaping the direction and outcomes of this study. Their unwavering commitment to my academic growth and professional development has been truly inspiring.
I am indebted to [Mentor’s Name] for their generous allocation of time and resources, their willingness to share their wealth of knowledge, and their unwavering dedication to pushing me to new heights. Their mentorship has not only enriched the quality of this research but has also had a profound impact on my personal and intellectual growth. I am truly fortunate to have had the privilege of working under their guidance.”
Sample 4: Acknowledgement for Institutional Support:
In this sample, we illustrate an acknowledgement that acknowledges the support and resources provided by institutions. It emphasizes the institutional backing that has facilitated the research process and contributed to its success.
“I would like to express my heartfelt gratitude to the faculty members and academic advisors who have provided guidance, feedback, and support throughout my academic journey. Their expertise, wisdom, and dedication to teaching and mentoring have been instrumental in shaping my research skills and scholarly pursuits.
I am grateful to [Name of Faculty Member/Advisor 1] for their unwavering support and invaluable insights. Their expertise and guidance have been critical in refining the research design, analyzing data, and interpreting findings. Their constructive feedback and intellectual discussions have truly enriched this study.
I would also like to acknowledge the contributions of [Name of Faculty Member/Advisor 2]. Their mentorship and encouragement have played a pivotal role in the development of my research abilities and have inspired me to reach for new heights. Their belief in my potential has been a constant source of motivation throughout this research journey.”
Sample 5: Acknowledgement for Peer Reviewers:
“I would like to express my deepest gratitude to the anonymous peer reviewers who have dedicated their time and expertise to provide valuable feedback and constructive criticism on this research paper. Their rigorous evaluation, insightful comments, and suggestions for improvement have immensely contributed to the quality and credibility of this work.
The meticulous review process conducted by the peer reviewers has helped shape and refine the content, methodology, and interpretation of this study. Their expertise in the field and their commitment to upholding scholarly standards have been crucial in ensuring the accuracy, validity, and relevance of the research findings.
I am sincerely grateful for the time and effort invested by each reviewer in thoroughly assessing this paper. Their detailed comments and recommendations have not only helped enhance the clarity and coherence of the manuscript but have also encouraged further reflection and refinement of the research.
The contributions of the peer reviewers are invaluable in the advancement of scientific knowledge and the improvement of academic publications. Their commitment to maintaining the rigor and integrity of the research process plays a pivotal role in fostering academic excellence and promoting the dissemination of high-quality research outcomes.”

When writing an acknowledgement in a research paper, begin by expressing gratitude to individuals, institutions, or organizations who have contributed to the research. Provide a sincere and concise acknowledgement, mentioning their specific contributions and the impact they made on the study.
While specific examples may vary depending on the research context, an acknowledgement section in a journal article typically acknowledges the contributions of individuals, funding sources, or institutions involved in the research process. It expresses gratitude for their support, guidance, or resources.
The purpose of the acknowledgement section in a research paper is to recognize and express gratitude to individuals or entities who have supported the research. It acknowledges their contributions, whether through funding, mentorship, technical assistance, data provision, or other forms of support.
When writing acknowledgements for a publication , start by identifying the key individuals or entities that have contributed to the research. Express gratitude for their support, mentioning specific contributions and the value they added to the study. Keep the acknowledgements concise and focused on the research context.
The acknowledgement section of a research paper should include acknowledgements for individuals or entities that have contributed to the research process. This may include mentors, advisors, funding agencies, research collaborators, or others who have provided valuable support, guidance, or resources.
While including an acknowledgement section in a research paper is not mandatory, it is a common practice in academic publishing. It provides an opportunity to acknowledge and appreciate the contributions of individuals or entities who have supported the research.
When writing an acknowledgements section for a literature review, acknowledge individuals or sources that have influenced and contributed to your understanding of the topic. Express gratitude for their insights, guidance, or resources that have shaped your literature review.
The terms “acknowledgement” and “acknowledgment” are both correct and interchangeable. The choice of spelling (with or without the “e”) may depend on regional or personal preferences.
To acknowledge a source in a research paper, use proper citation and referencing techniques according to the specific citation style guidelines. Include in-text citations and a corresponding entry in the reference list or bibliography to give credit to the original source.
Yes, you can acknowledge individuals who provided personal support in the acknowledgement section, such as family, friends, or loved ones. Recognize their emotional support, encouragement, or understanding during the research process.
Writing acknowledgements for a research paper allows you to express gratitude and acknowledge the invaluable contributions of individuals and institutions who have supported your work. By following the tips and utilizing the sample acknowledgements provided in this article, you can effectively and sincerely express your appreciation. Remember, acknowledgements are an opportunity to show your gratitude and give credit where it is due.
Acknowledgement for Paper Publication (10 Samples)
Acknowledgement for internship report: 10 samples and tips, leave a comment cancel reply.
Save my name, email, and website in this browser for the next time I comment.

How to write the Acknowledgements section of a research paper
References & Acknowledgements
Yateendra Joshi

Among all the sections of a typical research paper, the acknowledgements section is the easiest to write—which is probably why most books on writing research papers tend to ignore it. Yet, acknowledgements can be politically tricky. By forgetting to acknowledge those whom you should have acknowledged, you risk offending them; but even those whom you have acknowledged in your paper can take offence at the manner in which this is done. At times, when the help received is substantial, it can be hard to decide whether you should acknowledge the support or offer authorship instead. Wish this were as simple as remembering that it is Acknowledgments without the ‘e’ in US English but Acknowledgements with the ‘e’ in UK English.
This article discusses the purpose of the acknowledgements section in a research paper and offers tips on who should be mentioned in it and how, who should be excluded, and how the section should be formatted.
Why acknowledge?
In academic writing, the time-honoured method of acknowledging people is to cite their work, but that does not apply here. And yet, it is only proper that you put on record – by means of an appropriate mention in the acknowledgements section – any help that you received in conducting your research, in writing about it and publishing it. By doing so, albeit indirectly, you make your work more credible: for instance, when you acknowledge the help you received from a statistician in designing your experiment and in analysing its results, you reassure journal editors and, more important, those who review your manuscript, about the experimental design and the analysis of results. Similarly, acknowledging the help you received from a copy editor shows that you have taken care of the language, style, and formatting.

Who should be acknowledged ?
Broadly, you should acknowledge those who helped you by going beyond their normal call of duty, especially those whose help proved crucial to your work or who provided expertise that you lacked. Such people may include some of your peers, your mentors (research supervisors or guides), and even your students. If you received funding, the fact should be acknowledged. Some funding agencies may have specific instructions about how their funding should be mentioned; if that is so, make sure that the form of acknowledgement is consistent with such instructions. You should also consider acknowledging any material or other resources made available to you free of charge. However, if such help is mentioned as part of a conflict-of-interest statement, it should not be repeated in the acknowledgements section.
Consider including reviewers, even if they are anonymous, if their suggestions have resulted in a substantially improved manuscript.
It is also advisable to have your phrasing approved by those mentioned in the acknowledgements, because such a mention may imply that they approve of the contents of the research paper.

Who should not be acknowledged ?
Unlike dedications and acknowledgement commonly found in books, acknowledgements in research papers do not feature parents, family members, or friends (unless of course they qualify on other grounds). Similarly, those who provide a service as part of their job (laboratory technicians, field assistants, and so on) are usually excluded. Heads of departments, directors of laboratories, and people in similar positions also should not be acknowledged routinely: include them only if they went out of their way to help you.
Phrasing the acknowledgements
In general, be factual and avoid going overboard. Something along the lines of “The authors thank John Smith for advice on experimental design and statistical analysis” should be fine. Courtesy titles (Mr, Ms, Dr, etc.) before the names are rarely used (but check your target journal), and job titles or designations are seldom given. Avoid such expressions as ‘kind help’, ‘eternally grateful’, and ‘greatly indebted to’. If the acknowledgement is specifically by one of the authors of the paper, it is customary to use only the initials, as in “JS thanks...”.
In terms of sequence, any intellectual contributions come first, followed by technical support, help in revising and writing; financial support is mentioned at the end.
Formatting the acknowledgements
As a rule of thumb, the acknowledgement section should be a single short paragraph of say half a dozen lines. Examine the target journal for the format: whether the heading appears on a separate line or run on (that is, the text follows the heading on the same line). Check also whether the heading is in bold or in italics. The headings in the main body of the paper may be numbered, but the acknowledgement section is not numbered. Do not use any special formatting within the paragraph.
Effectively, acknowledgements signal the end of the main body of a research paper. Of course, they are followed by references—but that is another story.
for this article
Published on: Dec 02, 2021
- Acknowledgements Section
- manuscript writing
You're looking to give wings to your academic career and publication journey. We like that!
Why don't we give you complete access! Create a free account and get unlimited access to all resources & a vibrant researcher community.
One click sign-in with your social accounts

Sign up via email
1536 visitors saw this today and 1210 signed up.
Subscribe to Manuscript Writing
Translate your research into a publication-worthy manuscript by understanding the nuances of academic writing. Subscribe and get curated reads that will help you write an excellent manuscript.
Confirm that you would also like to sign up for free personalized email coaching for this stage.
Related Reading

Getting the references right: Citing social media sources

Style & Format
Handling abbreviations of journal names in references

Getting the references right: citing books as a source of information
How to write the Acknowledgements section of a research paper 4 min read
What you MUST know about plagiarism 7 min read
Who should be included in the acknowledgements section: A case study 4 min read
"Why should I acknowledge writing assistance if I have paid for it?" A case study 3 min read
Ethical declarations that authors should provide at the journal submission stage 10 min read
Trending Searches
- Statement of the problem
- Background of study
- Scope of the study
- Types of qualitative research
- Rationale of the study
- Concept paper
- Literature review
- Introduction in research
- Under "Editor Evaluation"
- Ethics in research
Recent Searches
- Review paper
- Responding to reviewer comments
- Predatory publishers
- Scope and delimitations
- Open access
- Plagiarism in research
- Journal selection tips
- Editor assigned
- Types of articles
- "Reject and Resubmit" status
- Decision in process
- Conflict of interest

Acknowledgement in Research Paper – A Quick Guide [5 Examples]
The acknowledgement section in your research paper is where you thank those who have helped or supported you throughout your research and writing. It is a short section of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
In this post, we are going to provide you with five examples of acknowlegdement section and a handful of best practices you can make your work look professional.

Saying thank you with style
How to write an acknowledgement: the complete guide for students, why should i include an acknowledgement in my research paper.
Acknowledging assistance and contributions from others can establish your integrity as a researcher. This will eventually make your work more credible.
What should be acknowledged about (aka thankful for)?
In your acknowledgement, you can show gratitude for those who provide you with resources in the following area:
- Technical help may include people who helped you by providing materials and supplies.
- Intellectual help includes academic advice and assistance.
- Mental help can be any kind of verbal support and encouragement.
- Financial support that is obviously related to monetary support
Who should be included in the acknowledgement of a research paper?
You can include everyone who helped you technically, intellectually, or financially (assistance with grants or monetary help) in the process of researching and writing your research paper. Except for your family and friends, you should always include the full names with the title of these individuals:
- Your profession, supervisor, or teacher
- Academic staff (e.g. lab assistant) of your school/college
- Your department, faculty, college, or school
- Classmates, teammates, co-workers, or colleague
- Friends and family members
You can start with your professor or the individuals who supported you the most throughout the research. And then you can continue by thanking your institution and then the reviewer who reviewed your paper. Then you can thank your friends and families and any other individual who helped.
What is the tone of the acknowledgement in a research paper?
You should write your acknowledgement in formal language with complete sentences. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
Note that personal pronouns such as ‘I, my, me …’ are nearly always used in the acknowledgements only. For the rest of the research paper, such personal pronouns are generally avoided.
Writing an acknowledgement for research paper is one of the important parts of your project report. You need to thank everyone for helping you with your paper . Here are some examples of acknowledgement for your research paper.
Acknowledgement in Research Paper: Example 1
Acknowledgement in research paper: example 2, acknowledgement in research paper: example 3, acknowledgement in research paper: example 4, acknowledgement in research paper: example 5.
You can use these or try to create your own version for your project report. Also, you can use our auto acknowledgement generator tool to automatically generate acknowledgement for your project.
Where should I put the acknowledgement section?
The acknowledgements section should appear between your title page and your introduction in your research paper.
How long is an acknowledgement in a research paper?
The acknowledgement section (usualy inserted as a page) of your research paper should consist of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
Should I use the full names of family members in an acknowledgement?
You do not necessarily need to use the full name for your family and friends (it would sound pretty awkward to use the full name of your parent or spouse right?), you should always include the full names with the title for all other individuals in your acknowledgement.
Can I use “first person” in an acknowledgement?
Yes. It is appropriate to write in the first person (‘I’ for a single author or ‘we’ for two or more).
What is an acknowledgement in academic writing?
An acknowledgement is a page is where you show appreciation to people who helped or supported you intellectually, mentally, or financially in your academic writing.
It should be no longer than one page.

More Definitions on Acknowledgement
- Acknowledgement vs Empathy What is the Difference Between Acknowledgement and Empathy?
- Acknowledgement vs Acceptance Does Acknowledgement Mean Acceptance? Lessons From History and the Bible
- Acknowledgement vs Agreement Is Acknowledgement the Same as Agreement?
- Acknowledgement Receipts vs Official Receipts What’s the Difference Between Acknowledgement Receipts and Official Receipts?
“Acknowledgement” vs “Acknowledgment”… …what the hack?

Both “acknowledgement” and “acknowledgment” are used in the English-speaking world. However, acknowledgement with the “e” in the middle is more commonly used. It is up to 24.5 times more popular in the top 5 English-speaking countries in the world.
Other Popular Acknowledgement Examples
For work or business Acknowledgement Receipt of Payment [4 Examples] Acknowledging Receipt of Documents: A Quick Guide with Examples Acknowledgement for Presentation [9 Examples] Acknowledgement for Job Offer [3 Examples] Acknowledgement for Business Plan [4 Examples] Acknowledgement for Work Immersion [5 Examples] Acknowledgement of Receipt of Appraisal [3 Examples] Acknowledegment of Debt [5 Examples] Resignation Acknowledgement for Employers [5 Examples]
Academic Acknowledgement for Research Paper [5 Examples] Acknowledgement for Internship Report [5 Examples] Acknowledgement for Thesis and Dissertation [15 Examples] Acknowledgement for Portfolio [5 Examples] Acknowledgement for Case Study [4 Examples] Acknowledgement for Academic Research Paper [5 Examples] Acknowledgement for College/School Assignment [5 Examples] Acknowledgemet to God in Reports [5 Examples]
Others Acknowledgement to Funeral Attendees [5 Examples] Funeral Acknowledgement Templates (for Newspapers and Websites) Common Website Disclaimers to Protect Your Online Business Notary Acknowledgement [5 Examples]
Acknowledgement Examples for School/College Projects
Most popular Acknowledgement For School/College Projects [7 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Project Class 11 and 12 Acknowledgement for Project of Class 8, 9 and 10 By subjects Acknowledgement for Accounting Project [3 Examples] Acknowledgement for Business Studies Project [5 Examples] Acknowledgement for Chemistry Project [5 Examples] Acknowledgement for Computer Project [5 Examples] Acknowledgement for Economics Project [5 Examples] Acknowledgement for English Project [5 Examples] Acknowledgement for Geography Project [5 Examples] Acknowledgement for History Project [5 Examples] Acknowledgement for Maths Project for Students [5 Examples] Acknowledgement for Physics Project [5 Examples] Acknowledgement for Social Science Project [5 Examples] Others Acknowledgement for Group Project [5 Examples] Acknowledgement for Graduation Project [5 Examples] Acknowledgement for Disaster Management Project [3 Examples] Acknowledgement for Yoga Project [3 Samples]
How-to Guides on Academic Writing and Others
Most popular How to Write an Acknowledgement: The Complete Guide for Students How to Write an Acknowledgement for College Project? How to Write a Dedication Page for a Thesis or Dissertation? More on acknowledgements How to Write Acknowledgment for a Dissertation or a Thesis? Is Acknowledgement and Dedication the Same? Thesis or Dissertation How to Write a Master’s Thesis: The Ultimate Guide How to Write a Thesis Proposal? How to Write an Abstract for a Thesis? How to Write a Preface for a Thesis? Others How to Write an Introduction for a Research Paper? 7 Real Research Paper Examples to Get You Started How to Write Cover Letter for an Internship Program? How to Write an Internship Acceptance Letter? How to Write a Leave Application? For Schools and the Workplace How to Write a Resignation Letter?
Introduction to Academic Writing
By O.P. Jindal Global University Duration: 16-hour Cost: FREE Gain an in-depth understanding of reading and writing as essential skills to conduct robust and critical research for your writing.
Writing in English at University
By Lund University Duration: 24-hour Cost: FREE Learn how to structure your text and arguments, quote sources, and incorporate editing and proofreading in your academic writing.
Academic English: Writing Specialization
By the University of California, Irvine Duration: 6 months Cost: Free 7-day trial, USD39 per month The skills taught in this Specialization will empower you to succeed in any college-level course or professional field. You’ll learn to conduct rigorous academic research and to express your ideas clearly in an academic format. Share your Course Certificates in your LinkedIn profile, on printed resumes, CVs, or other documents.

Thank you, your samples really helped me.
This is great! Your samples really helped me in my research. Thank you and more power!
I’m so grateful that you make this kind of blog, I really need this for my research. Thank you so much… God bless you.
Thank you so much for the samples you have provided, it has helped me a lot.
Leave a Comment Cancel Reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, generate accurate citations for free.
- Knowledge Base
- Dissertation
- Thesis & Dissertation Acknowledgements | Tips & Examples
Thesis & Dissertation Acknowledgements | Tips & Examples
Published on May 3, 2022 by Tegan George . Revised on July 18, 2023.
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process.
Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page.
In your acknowledgements, it’s okay to use a more informal style than is usually permitted in academic writing , as well as first-person pronouns . Acknowledgements are not considered part of the academic work itself, but rather your chance to write something more personal.
To get started, download our step-by-step template in the format of your choice below. We’ve also included sample sentence starters to help you construct your acknowledgments section from scratch.
Download Word doc Download Google doc
Instantly correct all language mistakes in your text
Upload your document to correct all your mistakes in minutes
Table of contents
Who to thank in your acknowledgements, how to write acknowledgements, acknowledgements section example, acknowledgements dos and don’ts, other interesting articles, frequently asked questions about the acknowledgements section.
Generally, there are two main categories of acknowledgements: professional and personal .
A good first step is to check your university’s guidelines, as they may have rules or preferences about the order, phrasing, or layout of acknowledgements. Some institutions prefer that you keep your acknowledgements strictly professional.
Regardless, it’s usually a good idea to place professional acknowledgements first, followed by any personal ones. You can then proceed by ranking who you’d like to thank from most formal to least.
- Chairs, supervisors, or defense committees
- Funding bodies
- Other academics (e.g., colleagues or cohort members)
- Editors or proofreaders
- Librarians, research/laboratory assistants, or study participants
- Family, friends, or pets
Typically, it’s only necessary to mention people who directly supported you during your thesis or dissertation. However, if you feel that someone like a high school physics teacher was a great inspiration on the path to your current research, feel free to include them as well.
Professional acknowledgements
It is crucial to avoid overlooking anyone who helped you professionally as you completed your thesis or dissertation. As a rule of thumb, anyone who directly contributed to your research process, from figuring out your dissertation topic to your final proofread, should be mentioned.
A few things to keep in mind include:
- Even if you feel your chair didn’t help you very much, you should still thank them first to avoid looking like you’re snubbing them.
- Be sure to follow academic conventions, using full names with titles where appropriate.
- If several members of a group or organization assisted you, mention the collective name only.
- Remember the ethical considerations around anonymized data. If you wish to protect someone’s privacy, use only their first name or a generic identifier (such as “the interviewees”)/
Personal acknowledgements
There is no need to mention every member of your family or friend group. However, if someone was particularly inspiring or supportive, you may wish to mention them specifically. Many people choose to thank parents, partners, children, friends, and even pets, but you can mention anyone who offered moral support or encouragement, or helped you in a tangible or intangible way.
Some students may wish to dedicate their dissertation to a deceased influential person in their personal life. In this case, it’s okay to mention them first, before any professional acknowledgements.
Prevent plagiarism. Run a free check.
After you’ve compiled a list of who you’d like to thank, you can then sort your list into rank order. Separate everyone you listed into “major thanks,” “big thanks,” and “minor thanks” categories.
- “Major thanks” are given to people who your project would be impossible without. These are often predominantly professional acknowledgements, such as your advisor, chair, and committee, as well as any funders.
- “Big thanks” are an in-between, for those who helped you along the way or helped you grow intellectually, such as classmates, peers, or librarians.
- “Minor thanks” can be a catch-all for everyone else, especially those who offered moral support or encouragement. This can include personal acknowledgements, such as parents, partners, children, friends, or even pets.
How to phrase your acknowledgements
To avoid acknowledgements that sound repetitive or dull, consider changing up your phrasing. Here are some examples of common sentence starters you can use for each category.
Note that you do not need to write any sort of conclusion or summary at the end. You can simply end the acknowledgements with your last thank you.
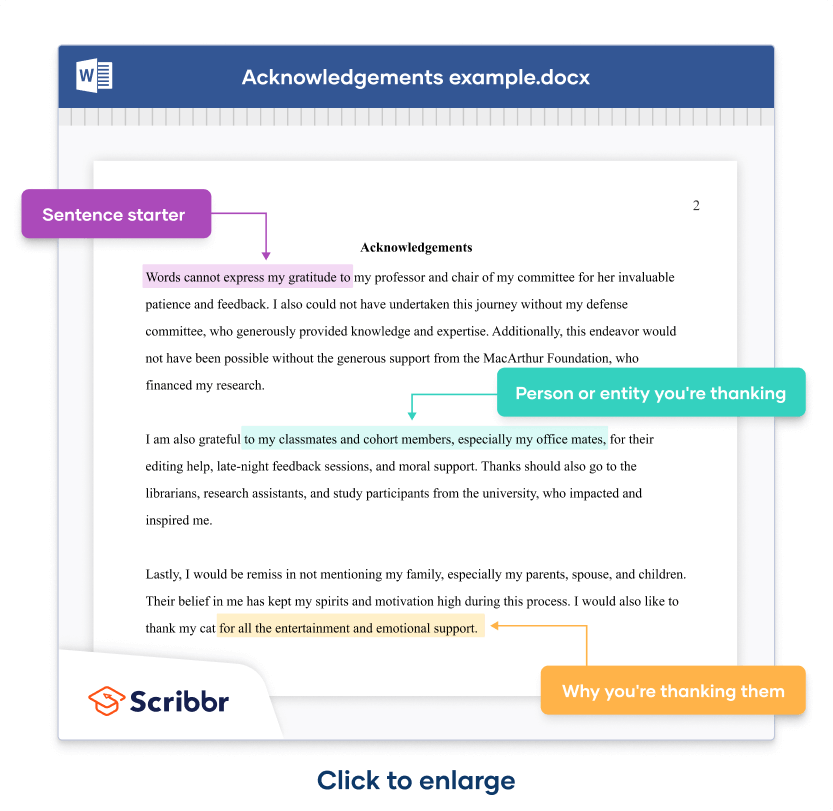
Here’s an example of how you can combine the different sentences to write your acknowledgements.
A simple construction consists of a sentence starter (in purple highlight ), followed by the person or entity mentioned (in green highlight ), followed by what you’re thanking them for (in yellow highlight .)
Acknowledgements
Words cannot express my gratitude to my professor and chair of my committee for her invaluable patience and feedback. I also could not have undertaken this journey without my defense committee, who generously provided knowledge and expertise. Additionally, this endeavor would not have been possible without the generous support from the MacArthur Foundation, who financed my research .
I am also grateful to my classmates and cohort members, especially my office mates, for their editing help, late-night feedback sessions, and moral support. Thanks should also go to the librarians, research assistants, and study participants from the university, who impacted and inspired me.
Lastly, I would be remiss in not mentioning my family, especially my parents, spouse, and children. Their belief in me has kept my spirits and motivation high during this process. I would also like to thank my cat for all the entertainment and emotional support.
- Write in first-person, professional language
- Thank your professional contacts first
- Include full names, titles, and roles of professional acknowledgements
- Include personal or intangible supporters, like friends, family, or even pets
- Mention funding bodies and what they funded
- Appropriately anonymize or group research participants or non-individual acknowledgments
Don’t:
- Use informal language or slang
- Go over one page in length
- Mention people who had only a peripheral or minor impact on your work
If you want to know more about AI for academic writing, AI tools, or research bias, make sure to check out some of our other articles with explanations and examples or go directly to our tools!
Research bias
- Anchoring bias
- Halo effect
- The Baader–Meinhof phenomenon
- The placebo effect
- Nonresponse bias
- Deep learning
- Generative AI
- Machine learning
- Reinforcement learning
- Supervised vs. unsupervised learning
(AI) Tools
- Grammar Checker
- Paraphrasing Tool
- Text Summarizer
- AI Detector
- Plagiarism Checker
- Citation Generator
In the acknowledgements of your thesis or dissertation, you should first thank those who helped you academically or professionally, such as your supervisor, funders, and other academics.
Then you can include personal thanks to friends, family members, or anyone else who supported you during the process.
Yes, it’s important to thank your supervisor(s) in the acknowledgements section of your thesis or dissertation .
Even if you feel your supervisor did not contribute greatly to the final product, you must acknowledge them, if only for a very brief thank you. If you do not include your supervisor, it may be seen as a snub.
The acknowledgements are generally included at the very beginning of your thesis , directly after the title page and before the abstract .
In a thesis or dissertation, the acknowledgements should usually be no longer than one page. There is no minimum length.
You may acknowledge God in your dissertation acknowledgements , but be sure to follow academic convention by also thanking the members of academia, as well as family, colleagues, and friends who helped you.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
George, T. (2023, July 18). Thesis & Dissertation Acknowledgements | Tips & Examples. Scribbr. Retrieved April 10, 2024, from https://www.scribbr.com/dissertation/acknowledgements/
Is this article helpful?
Tegan George
Other students also liked, dissertation layout and formatting, thesis & dissertation title page | free templates & examples, how to write an abstract | steps & examples, "i thought ai proofreading was useless but..".
I've been using Scribbr for years now and I know it's a service that won't disappoint. It does a good job spotting mistakes”
The Acknowledgements Section
How to write the acknowledgements for your thesis or dissertation
By: Derek Jansen (MBA) | Reviewers: Dr Eunice Rautenbach | January 2024
Writing the acknowledgements section of your thesis might seem straightforward, but it’s more than just a list of names . In this post, we’ll unpack everything you need to know to write up a rock-solid acknowledgements section for your dissertation or thesis.
Overview: The Acknowledgements
- What (exactly) is the acknowledgements section?
Who should you acknowledge?
- How to write the section
- Practical example
- Free acknowledgements template
- Key takeaways
What is the acknowledgements section?
The acknowledgements section of your thesis or dissertation is where you give thanks to the people who contributed to your project’s success. Generally speaking, this is a relatively brief, less formal section.
With the acknowledgements section, you have the opportunity to show appreciation for the guidance, support, and resources provided by others during your research journey. We’ll unpack the exact contents, order and structure of this section in this post.
Need a helping hand?
Although this is a less “academic” section, acknowledging the right people in the correct order is still important. Typically, you’ll start with the most formal (academic) support received, before moving on to other types of support.
Here’s a suggested order that you can follow when writing up your acknowledgements:
Level 1: Supervisors and academic staff
Start with those who have provided you with academic guidance, including your supervisor, advisors, and other faculty members.
Level 2: Funding bodies or sponsors
If your research was funded, acknowledging these organisations is essential. You don’t need to get into the specifics of the funding, but you should recognise the important role that this made in bringing your project to life.
Level 3: Colleagues and peers
Next you’ll want to mention those who contributed intellectually to your work, including your fellow cohort members and researchers.
Level 4: Family, friends and pets
Last but certainly not least, you should acknowledge your personal (non-academic) support system – those who have provided emotional and moral support. If Fido kept you company during those long nights hunched over the keyboard, you can also thank him here 🙂
As you can see, the order of the acknowledgements goes from the most academic to the least . Importantly, your thesis or dissertation supervisor (sometimes also called an advisor) generally comes first . This is because they are typically the person most involved in shaping your project (or at least, they should be). Plus, they’re oftentimes involved in marking your final work and so a kind word never hurts…
All that said, remember that your acknowledgements section is personal . So, feel free to adjust this order, but do pay close attention to any guidelines or rules provided by your university. If they specify a certain order or set of contents, follow their instructions to the letter.

How to write the acknowledgements section
In terms of style, try to strike a balance between conveying a formal tone and a personal touch . In practical terms, this means that you should use plain, straightforward language (this isn’t the time for heavy academic jargon), but avoid using any slang, nicknames, etc.
As a guide, you’ll typically use some of the following phrases in the acknowledgements section:
I would like to express my appreciation to… for their help with… I’m particularly grateful to… as they provided… I could not have completed this project without… as this allowed me to… Special thanks to… who did… I had the pleasure of working with… who helped me… I’d also like to recognise… who assisted me with…
In terms of positioning, the acknowledgements section is typically in the preliminary matter , most commonly after the abstract and before the table of contents. In terms of length, this section usually spans one to three paragraphs , but there’s no strict word limit (unless your university’s brief states otherwise, of course).
If you’re unsure where to place your acknowledgements or what length to make this section, it’s a good idea to have a look at past dissertations and theses from your university and/or department to get a clearer view of what the norms are.

Practical Example
Alright, let’s look at an example to give you a better idea of what this section looks like in practice.
I would like to express my deepest gratitude to Professor Smith, whose expertise and knowledge were invaluable during this research. My sincere thanks also go to the University Research Fund for their financial support. I am deeply thankful to my colleagues, John and Jane, for their insightful discussions and moral support. Lastly, I must acknowledge my family for their unwavering love and encouragement. Without your support, this project would not have been possible.
As you can see in this example, the section is short and to the point , working from formal support through to personal support.
To simplify the process, we’ve created a free template for the acknowledgements section. If you’re interested, you can download a copy here .

FAQs: Acknowledgements
Can i include some humour in my acknowledgements.
A touch of light humour is okay, but keep it appropriate and professional. Remember that this is still part of an academic document.
Can I acknowledge someone who provided informal or emotional support?
Yes, you can thank anyone who offered emotional support, motivation, or even informal advice that helped you during your studies. This can include friends, family members, or a mentor/coach who provided guidance outside of an academic setting.
Should I mention any challenges or difficulties I faced during my research?
While the acknowledgements section is primarily for expressing gratitude, briefly mentioning significant challenges you overcame can highlight the importance of the support you received. That said, you’ll want to keep the focus on the gratitude aspect and avoid delving too deeply into the challenges themselves.
Can I acknowledge the contribution of participants in my research?
Absolutely. If your research involved participants, especially in fields like social sciences or human studies, acknowledging their contribution is not only courteous but also an ethical practice. It shows respect for their participation and contribution to your research.
How do I acknowledge posthumous gratitude, for someone who passed away during my study period?
Acknowledging a deceased individual who played a significant role in your academic journey can be done respectfully. Mention them in the same way you would a living contributor, perhaps adding a note of remembrance.
For example, “I would like to posthumously acknowledge John McAnders for their invaluable advice and support in the early stages of this research.”.
Is there a limit to the number of people I can acknowledge?
How do i acknowledge a group or organisation.
When thanking a group or organization, mention the entity by name and, if applicable, include specific individuals within the organization who were particularly helpful.
For example, “I extend my thanks to The Speakers Foundation for their support, particularly Mr Joe Wilkins, for their guidance.”
Recap: Key Takeaways
Writing the acknowledgements section of your thesis or dissertation is an opportunity to express gratitude to everyone who helped you along the way.
Remember to:
- Acknowledge those people who significantly contributed to your research journey
- Order your thanks from formal support to personal support
- Maintain a balance between formal and personal tones
- Keep it concise
In a nutshell, use this section to reflect your appreciation in a genuinely and professionally way.

Psst… there’s more (for free)
This post is part of our dissertation mini-course, which covers everything you need to get started with your dissertation, thesis or research project.
You Might Also Like:

Submit a Comment Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
- Print Friendly
- Resources Home 🏠
- Try SciSpace Copilot
- Search research papers
- Add Copilot Extension
- Try AI Detector
- Try Paraphraser
- Try Citation Generator
- April Papers
- June Papers
- July Papers

How to Write Acknowledgement in Research Paper

Table of Contents
Writing an acknowledgement in a research paper is an integral part of the process. It is a formal way of expressing gratitude to the individuals and institutions that contributed to the completion of your research.
This section, though not mandatory, holds significant value as it acknowledges the efforts of those who assisted you in the successful completion of your project. In this comprehensive guide, we will delve into the intricacies of writing an effective acknowledgement for your research paper.
Introduction
Acknowledgements serve a crucial role in research papers . They not only express gratitude but also provide a sense of credibility to your work. Acknowledging the contributions of others shows that your research is a collective effort, which can enhance the perceived validity of your findings.
Moreover, acknowledgements can also serve as a platform for you to demonstrate your professional courtesy and respect for the individuals and institutions that have supported your research. This can help in fostering positive relationships, which can be beneficial for your future research endeavors.
Who to acknowledge in your research paper ?
Deciding who to acknowledge in your research paper can be a challenging task. It is important to ensure that you do not overlook anyone who has contributed to your research. Here are some categories of individuals and institutions that you might consider acknowledging:
Academic advisors and supervisors:
Your academic advisors and supervisors are likely to be your first point of contact for guidance and support during your research. They provide valuable insights, feedback, and direction, which can significantly influence the outcome of your research.
Therefore, acknowledging them in your research paper is a way of expressing your gratitude for their assistance and guidance. It also shows your respect for their expertise and dedication to your research.
Research participants and collaborators
Research participants and collaborators play a crucial role in the success of your research. They provide the data or information necessary for your research, making their contribution invaluable.
Acknowledging them in your research paper is a way of showing your appreciation for their time and effort. It also symbolizes your respect for their contribution to your research.
Funding bodies and institutions:
Funding bodies and institutions provide the financial support necessary for conducting your research. Without their support, it might be challenging to carry out your research effectively.
Therefore, acknowledging them in your research paper is a way of expressing your gratitude for their financial support. It also shows your appreciation for their trust in your research capabilities.
How to write acknowledgements for your research paper?
Writing acknowledgements for your research paper involves more than simply listing names. It requires a thoughtful and sincere expression of gratitude. Here are some steps to guide you in writing effective acknowledgements:
Start with the most significant contributions:
Begin your acknowledgements by expressing gratitude to those who have made the most significant contributions to your research. This could be your academic advisors, supervisors, or funding bodies. Starting with the most significant contributions helps to set the tone for the rest of your acknowledgements.
Ensure that you express your gratitude sincerely and professionally. Avoid using overly emotional or informal language as this can undermine the professionalism of your acknowledgements.
Acknowledge other contributors:
After acknowledging the most significant contributors, proceed to acknowledge other individuals and institutions that have supported your research. This could include research participants, collaborators, and other supportive individuals or institutions.
When acknowledging these contributors, be sure to express your gratitude sincerely and professionally. Also, ensure that you acknowledge each contributor individually to show your appreciation for their unique contribution.
Use appropriate language and tone:
The language and tone you use in your acknowledgements can significantly influence how they are perceived. Therefore, it is important to use appropriate language and maintain a professional tone throughout your acknowledgements.
Use formal language and avoid using jargon or colloquial expressions. Also, maintain a consistent tone throughout your acknowledgements to ensure that they are coherent and easy to read.
Examples of acknowledgements in research papers
Here are a few examples that demonstrate how to acknowledge different contributors effectively:
"I would like to express my deepest gratitude to my advisor, Professor ABC, for his invaluable guidance and support throughout this research. His expertise and dedication have been a source of inspiration and motivation."
Research participants and collaborators:
"I am deeply grateful to all the participants who generously shared their time and experiences for this research. Their contributions have been instrumental in the success of this study."
"This research was made possible by the generous funding from ABC Foundation. I am profoundly grateful for their support and trust in my research capabilities."
Writing acknowledgements in a research paper is a thoughtful process that requires careful consideration of who to acknowledge and how to express gratitude. By following the guidelines and examples provided in this article, you can write effective acknowledgements that reflect your appreciation and respect for the contributions of others to your research.
Remember, acknowledgements are more than just a formality. They are an opportunity to express your gratitude and respect for the individuals and institutions that have supported your research journey. So, take the time to write acknowledgements that are sincere, professional, and reflective of your gratitude.
You might also like

AI for Meta Analysis — A Comprehensive Guide

How To Write An Argumentative Essay

Beyond Google Scholar: Why SciSpace is the best alternative
Reference management. Clean and simple.
Dissertation acknowledgments [with examples]

What are dissertation acknowledgements?
What to consider when writing your dissertation acknowledgments, who to thank in your dissertation acknowledgments, what (and what not) to write in your dissertation acknowledgments, good examples of dissertation acknowledgments, a final word on writing dissertation acknowledgments: have fun, frequently asked questions about dissertation acknowledgments, related articles.
While you may be the sole author of your dissertation, there are lots of people who help you through the process—from your formal dissertation advisors to the friends who may have cooked meals so that you could finish your last chapter . Dissertation acknowledgments are a chance to thank everyone who had a hand in the completion of your project.
Dissertation acknowledgments are a brief statement of your gratitude to advisors, professors, peers, family, and friends for their help and expertise.
In this guide, we’ll cover:
- the most important things to consider when you’re writing your dissertation acknowledgments
- who to thank in your dissertation acknowledgments
- what (and what not) to write in your dissertation acknowledgments
- short examples of dissertation acknowledgments
Once you’re at the stage where you’re writing your dissertation acknowledgments, you may be tempted to kick back and relax. After all, the hard part of writing the dissertation itself is over and a list of thanks should be simple to churn out.
However, the acknowledgments are an important part of your overall work and are something that most people who read your dissertation, including prospective employers, will look at.
Tip: The best dissertation acknowledgements are concise, sincere, and memorable.
Approach this part of the process, brief as it may be compared to the long haul of writing the dissertation, with the same high level of care and attention to detail. It’s an explicit and permanent statement of who made a real impact on your work and contributed to your academic success.
Plus, the people you thank are often deeply moved by being included—some even go so far as to frame the acknowledgments. Aim to make yours sincere, memorable and something that people will be touched by.
First things first: who should you include in your dissertation acknowledgments? If you’re not sure who to thank, try the brainstorming technique to generate some ideas. Consider these two approaches:
- Make a list of everyone, both professional and personal, who was involved at any point during your work on your dissertation, and then thin down the list from there.
- Make a list of the pivotal aspects of your process and think about who was involved and how they helped.
As you select the people and groups to include in your dissertation acknowledgments, keep in mind that it’s essential to acknowledge your supervisor and anyone else with a visible connection to your work.
It’s an unfortunate reality that not every supervisor goes above and beyond to provide feedback and guidance to the students they are supposed to supervise. However, leaving them out, even if you personally felt disappointed by their involvement or lack thereof, could be seen as a snub.
You should end up with a fairly short list of people to thank. While being mindful of professional etiquette and personal feelings, be choosy about who makes the final cut since your acknowledgments should be limited to no more than a page.
Now that you have your list of people and groups to thank, it’s time to start writing. Before your first pen or keystroke, however, check your university’s guidelines as your institution may have specific rules around what can and cannot be included.
The standard practice is to begin with the formal and then progress to the informal, so the first people to mention would be:
- supervisors
- committee members
- other professional contacts
Use their full names and titles and go into brief detail about how they contributed to your work.
Once those are done, you can move on to the personal thanks, which can include friends, family, even pets. If you are so inclined, it is also considered appropriate to thank God or make mention of spiritual support.
You may also choose to inject a little humor at this point, but don’t get carried away and definitely don’t include sarcasm or critical comments of any kind, including self-critical ones. Remember that the acknowledgments precede your dissertation, so you want to be taken seriously.
A couple more basics that are essential when creating your acknowledgments:
- Position: Acknowledgments should be placed after the title page and before the abstract.
- Perspective: Write from the first-person perspective and speak in your own voice.
A really good way to get a sense of how to write your own dissertation acknowledgments is to read ones written by others. Notice which ones you respond particularly well to and use them as a model upon which to base your own.
Here are some good examples to help you get started:
I couldn’t have reached this goal without the help of many people in my life. I’d like to take this opportunity to thank them for their support.
First, my sincere thanks to my dissertation committee. The value of their guidance cannot be overstated. Dr. Elaine Gooding and Dr. Matthew Hunter provided much wisdom that helped me chart my course. I couldn’t have asked for a better supervisor than Dr. Fiona Moore, whose knowledge and experience guided me every step of the way.
Next, I’d like to thank my partner, Elliott. Your votes of confidence kept me going when my spirits dipped. I couldn’t have done this without you.
Last but not least, I’d like to acknowledge the emotional support provided by my family and friends. We made it to the top of the mountain! I look forward to celebrating with all of you.
This example is shorter, but still contains the key components:
Several people played a decisive role in my success and I would like to take this opportunity to thank them.
My chair, Dr. Ronald Saulk, provided invaluable support and infinite patience and I am truly grateful for all of his wisdom and guidance. I also owe the entire staff of the Wilhelm Library a debt of gratitude. From tracking down books and arranging for interlibrary loans to keeping the coffee maker in the lobby well-stocked and in good working order, they offered the practical help and kind gestures that made all the difference.
I’d also like to thank my family and God, for always being there for me.
One final piece of advice: enjoy this process. Writing a dissertation doesn’t happen every day, and the opportunity to acknowledge the important people in your life in a published format is as rare as it is wonderful.
What’s more, this part of your dissertation is unlike any other. It’s unbounded by the conventions that apply to the formal work. It’s a chance to really flex some creative muscle and let your personality shine through. So make the most of it and have fun!
In your dissertation acknowledgments, you thank everyone who has contributed to your work or supported you along the way. Who you want to thank is a very personal choice, but you should include your supervisors and anyone else with a visible connection to your work. You may also thank friends, family, and partners.
First, you need to come up with a list of people you want to thank in your dissertation acknowledgments. As a next step, begin with the formal and then progress to the informal, so the first people to mention would be supervisors, mentors, committees, and other professional contacts. Then, you can move on to the personal thanks, which can include friends, family, even pets.
Who you acknowledge in your dissertation is ultimately up to you. You should, however, thank your supervisor and anyone else with a visible connection to your work. Leaving them out, even if you personally felt disappointed by their involvement or lack thereof, could be seen as a snub. In addition, you can thank friends, partners or family.
There are many ways so you can acknowledge your dissertation supervisor. Some examples can be found in this article above. If you need more examples, you can find them here .
While acknowledgments are usually more present in academic theses, they can also be a part of research papers. In academic theses, acknowledgments are usually found at the beginning, somewhere between abstract and introduction. In research papers, acknowledgments are usually found at the end of the paper.
- SpringerLink shop
Acknowledgments and References
Acknowledgments.
This usually follows the Discussion and Conclusions sections. Its purpose is to thank all of the people who helped with the research but did not qualify for authorship (check the target journal’s Instructions for Authors for authorship guidelines). Acknowledge anyone who provided intellectual assistance, technical help (including with writing and editing), or special equipment or materials.
TIP: The International Committee of Medical Journal Editors has detailed guidelines on who to list as an author and who to include in the Acknowledgments that are useful for scientists in all fields.
Some journals request that you use this section to provide information about funding by including specific grant numbers and titles. Check your target journal’s instruction for authors for specific instructions. If you need to include funding information, list the name(s) of the funding organization(s) in full, and identify which authors received funding for what.
As references have an important role in many parts of a manuscript, failure to sufficiently cite other work can reduce your chances of being published. Every statement of fact or description of previous findings requires a supporting reference.
TIP: Be sure to cite publications whose results disagree with yours. Not citing conflicting work will make readers wonder whether you are really familiar with the research literature. Citing conflicting work is also a chance to explain why you think your results are different.
It is also important to be concise. You need to meet all the above needs without overwhelming the reader with too many references—only the most relevant and recent articles need to be cited. There is no correct number of references for a manuscript, but be sure to check the journal’s guidelines to see whether it has limits on numbers of references.
TIP: Never cite a publication based on what you have read in a different publication (such as a review), or based only on the publication’s abstract. These may mislead you and readers. Read the publication itself before you cite it, and then check the accuracy of the citation again before submitting your manuscript.
You should reference other work to:
- Establish the origin of ideas
When you refer to an idea or theory, it is important to let your readers know which researcher(s) came up with the idea. By citing publications that have influenced your own work, you give credit to the authors and help others evaluate the importance of particular publications. Acknowledging others’ contributions is also an important ethical principle.
- Justify claims
In a scientific manuscript, all statements must be supported with evidence. This evidence can come from the results of the current research, common knowledge, or from previous publications. A citation after a claim makes it clear which previous study supports the claim.
- Provide a context for your work
By highlighting related works, citations help show how a manuscript fits into the bigger picture of scientific research. When readers understand what previous studies found and what puzzles or controversies your study relates to, they will better understand the meaning of your work.
- Show there is interest your field of research
Citations show that other researchers are performing work similar to your own. Having current citations will help journal editors see that there is a potential audience for your manuscript.
Back │ Next

- Acknowledgements for PhD Thesis and Dissertations – Explained
- Doing a PhD
The Purpose of Acknowledgements
The acknowledgement section of a thesis or dissertation is where you recognise and thank those who supported you during your PhD. This can be but is not limited to individuals, institutions or organisations.
Although your acknowledgements will not be used to evaluate your work, it is still an important section of your thesis. This is because it can have a positive (or negative for that matter) influence the perception of your reader before they even reach the main body of your work.
Who Should I Acknowledge?
Acknowledgements for a PhD thesis will typically fall into one of two categories – professional or personal.
Within these categories, who you thank will ultimately be your decision. However, it’s imperative that you pay special attention to the ‘professional’ group. This is because not thanking someone who has played an important role in your studies, whether it be intentional or accidental, will more often than not be seen as a dismissal of their efforts. Not only would this be unfair if they genuinely helped you, but from a certain political aspect, it could also jeopardise any opportunities for future collaborations .
Professional Acknowledgements
This may include, but is not limited to:
- Funding bodies/sponsorship providers
- Supervisors
- Research group and lab assistants
- Research participants
- Proofreaders
Personal Acknowledgements
- Key family members and friends
- Individuals who inspired you or directly influenced your academic journey
- Anyone else who has provided personal support that you would like to mention
It should be noted that certain universities have policies which state only those who have directly supported your work, such as supervisors and professors, should be included in your acknowledgements. Therefore, we strongly recommend that you read your university guidelines before writing this section of your thesis.
How to Write Acknowledgements for PhD Thesis
When producing this section, your writing style can be more informal compared to the rest of your thesis. This includes writing in first person and using more emotive language. Although in most cases you will have complete freedom in how you write this section of your thesis, it is still highly advisable to keep it professional. As mentioned earlier, this is largely because it will be one of the first things your assessors will read, and so it will help set the tone for the rest of your work.
In terms of its structure, acknowledgements are expected to be ordered in a manner that first recognises the most formal support before moving onto the less formal support. In most cases, this follows the same order that we have outlined in the ‘Who Should I Thank’ section.
When thanking professionals, always write out their full name and provide their title. This is because although you may be on a first-name basis with them, those who read your thesis will not. By providing full names and titles, not only do you help ensure clarity, but it could also indirectly contribute to the credibility of your thesis should the individual you’re thanking be well known within your field.
If you intend to include a list of people from one institution or organisation, it is best to list their names in alphabetical order. The exception to this is when a particular individual has been of significant assistance; here, it would be advisable to list them.
How Long Should My Acknowledgements Be?
Acknowledgements vary considerably in length. Some are a single paragraph whilst some continue for up to three pages. The length of your acknowledgement page will mostly depend on the number of individuals you want to recognise.
As a general rule, try to keep your acknowledgements section to a single page. Although there are no word limits, creating a lengthy acknowledgements section dilutes the gratitude you’re trying to express, especially to those who have supported you the most.
Where Should My Acknowledgements Go?
In the vast majority of cases, your acknowledgements should appear directly after your abstract and before your table of contents.
However, we highly advise you to check your university guidelines as a few universities set out their own specific order which they will expect you to follow.
Phrases to Help You Get Started

We appreciate how difficult it can be to truly show how grateful you are to those who have supported you over the years, especially in words.
To help you get started, we’ve provided you with a few examples of sentences that you can complete or draw ideas from.
- I am deeply grateful to XXX…
- I would like to express my sincere gratitude to XXX…
- I would like to offer my special thanks to XXX…
- I would like to extend my sincere thanks to XXX…
- …for their assistance at every stage of the research project.
- …for their insightful comments and suggestions.
- …for their contribution to XXX.
- …for their unwavering support and belief in me.
Thesis Acknowledgement Examples
Below are three PhD thesis acknowledgment samples from which you can draw inspiration. It should be noted that the following have been extracted from theses which are freely available in the public domain. Irrespective of this, references to any individual, department or university have been removed for the sake of privacy.
First and foremost I am extremely grateful to my supervisors, Prof. XXX and Dr. XXX for their invaluable advice, continuous support, and patience during my PhD study. Their immense knowledge and plentiful experience have encouraged me in all the time of my academic research and daily life. I would also like to thank Dr. XXX and Dr. XXX for their technical support on my study. I would like to thank all the members in the XXX. It is their kind help and support that have made my study and life in the UK a wonderful time. Finally, I would like to express my gratitude to my parents, my wife and my children. Without their tremendous understanding and encouragement in the past few years, it would be impossible for me to complete my study.
I would like to thank my supervisors Dr. XXX and Dr. XXX for all their help and advice with this PhD. I would also like to thank my sisters, whom without this would have not been possible. I also appreciate all the support I received from the rest of my family. Lastly, I would like to thank the XXX for the studentship that allowed me to conduct this thesis.
I would like to thank my esteemed supervisor – Dr. XXX for his invaluable supervision, support and tutelage during the course of my PhD degree. My gratitude extends to the Faculty of XXX for the funding opportunity to undertake my studies at the Department of XXX, University of XXX. Additionally, I would like to express gratitude to Dr. XXX for her treasured support which was really influential in shaping my experiment methods and critiquing my results. I also thank Dr. XXX, Dr. XXX, Dr. XXX for their mentorship. I would like to thank my friends, lab mates, colleagues and research team – XXX, XXX, XXX, XXX for a cherished time spent together in the lab, and in social settings. My appreciation also goes out to my family and friends for their encouragement and support all through my studies.
Browse PhDs Now
Join thousands of students.
Join thousands of other students and stay up to date with the latest PhD programmes, funding opportunities and advice.

How to Draft the Acknowledgment Section of a Manuscript
What is the Purpose of the Acknowledgements Section in a Research Paper?
The acknowledgment section is an integral part of all academic research papers. It provides appropriate recognition to all contributors for their hard work. We discuss here, the relevant guidelines for acknowledging contributors.
Defining Who Is Acknowledged
The acknowledgment section helps identify the contributors responsible for specific parts of the project. It can include:
- Non-authors (colleagues, friends, supervisor, etc.)
- Funding sources
- Editing services ,
- Administrative staff
In academic writing, the information presented in the acknowledgment section should be kept brief. It should only mention people directly involved with the project. In other words, one should not consider thanking ones’ parents for moral and financial support.
Acknowledging contributors is necessary. However, you must know the difference between an author and a contributor . The International Committee of Medical Journal Editors ( ICMJE ) defines four criteria to assign authorship.
He or she has to have
- Made substantial conceptual or design contributions or gathered and analyzed important data, and
- Either helped draft or critically revise the paper in keeping with important intellectual content, and
- Provided final approval before publishing, and
- Agreed to be accountable for the accuracy of the work
These authors and their affiliations will be listed at the beginning of the paper. The “corresponding author” will also be listed a second time and will directly correspond with the journal to ensure documentation requirements are met.
Many journals now ask that you provide the role of each author in your acknowledgment section. For example, a typical statement of authors’ contributions might be as follows (note that only last names are used unless ambiguous):
Smith conducted the data analysis and created the tables and figures. Jones provided his technological expertise for GIS tracking. Johnson provided a factual review and helped edit the manuscript.
This type of acknowledgment provides your reader with a good sense of who was responsible for each part of your research and manuscript.
Non-Author Contributors
There are many people involved in a research project who are not authors but have provided valuable contributions. For example, one person’s responsibility might be to seek project funding; another’s might be to supervise laboratory staff. A few others might have provided valuable services such as technical editing and writing or offering help in reviewing and revising the manuscript for grammar and syntax. These people should also be mentioned in the acknowledgment section of your manuscript.
Acknowledgment should also be provided for writing assistance, technical editing, language editing, and proofreading . Therefore, editing companies need to be duly acknowledged in professionally edited manuscripts as per the ICMJE guidelines.
It is necessary to acknowledge editing companies in professionally edited manuscripts, even though these companies are paid for their work.
Acknowledgment Format
Unlike the main body of your paper, the format for your acknowledgment section can be more personal. It is permissible to use personal pronouns in this section. For example,
I thank the following individuals for their expertise and assistance throughout all aspects of our study and for their help in writing the manuscript.
Keep in mind that many guidelines indicate that funding sources be listed separately from the acknowledgment section. In addition, the sources (funding agencies) might have specific guidelines that you must follow. Please be sure to comply with these sources and your author guidelines.
For more information on authors and contributors , read articles on the Enago Academy website.
What types and formats of acknowledgments have you incorporated into your manuscripts? Please share your thoughts in the comments section below. Do you need help with manuscript editing ? Make sure you visit enago.com today!
Thanks!!! This information helped me a lot in finishing my research paper.
This information was very useful for preparing this paper.
We would like to thank Enago (www.enago.com) for the English language review.”
Rate this article Cancel Reply
Your email address will not be published.

Enago Academy's Most Popular Articles

- AI in Academia
- Infographic
- Manuscripts & Grants
- Reporting Research
- Trending Now
Can AI Tools Prepare a Research Manuscript From Scratch? — A comprehensive guide
As technology continues to advance, the question of whether artificial intelligence (AI) tools can prepare…

Abstract Vs. Introduction — Do you know the difference?
Ross wants to publish his research. Feeling positive about his research outcomes, he begins to…

- Old Webinars
- Webinar Mobile App
Demystifying Research Methodology With Field Experts
Choosing research methodology Research design and methodology Evidence-based research approach How RAxter can assist researchers

- Manuscript Preparation
- Publishing Research
How to Choose Best Research Methodology for Your Study
Successful research conduction requires proper planning and execution. While there are multiple reasons and aspects…

Top 5 Key Differences Between Methods and Methodology
While burning the midnight oil during literature review, most researchers do not realize that the…
Discussion Vs. Conclusion: Know the Difference Before Drafting Manuscripts
Annex Vs. Appendix: Do You Know the Difference?

Sign-up to read more
Subscribe for free to get unrestricted access to all our resources on research writing and academic publishing including:
- 2000+ blog articles
- 50+ Webinars
- 10+ Expert podcasts
- 50+ Infographics
- 10+ Checklists
- Research Guides
We hate spam too. We promise to protect your privacy and never spam you.
I am looking for Editing/ Proofreading services for my manuscript Tentative date of next journal submission:

What should universities' stance be on AI tools in research and academic writing?
- Discoveries
- Right Journal
- Journal Metrics
- Journal Fit
- Abbreviation
- In-Text Citations
- Bibliographies
- Writing an Article
- Peer Review Types
- Acknowledgements
- Withdrawing a Paper
- Form Letter
- ISO, ANSI, CFR
- Google Scholar
- Journal Manuscript Editing
- Research Manuscript Editing
Book Editing
- Manuscript Editing Services
Medical Editing
- Bioscience Editing
- Physical Science Editing
- PhD Thesis Editing Services
- PhD Editing
- Master’s Proofreading
- Bachelor’s Editing
- Dissertation Proofreading Services
- Best Dissertation Proofreaders
- Masters Dissertation Proofreading
- PhD Proofreaders
- Proofreading PhD Thesis Price
- Journal Article Editing
- Book Editing Service
- Editing and Proofreading Services
- Research Paper Editing
- Medical Manuscript Editing
- Academic Editing
- Social Sciences Editing
- Academic Proofreading
- PhD Theses Editing
- Dissertation Proofreading
- Proofreading Rates UK
- Medical Proofreading
- PhD Proofreading Services UK
- Academic Proofreading Services UK
Medical Editing Services
- Life Science Editing
- Biomedical Editing
- Environmental Science Editing
- Pharmaceutical Science Editing
- Economics Editing
- Psychology Editing
- Sociology Editing
- Archaeology Editing
- History Paper Editing
- Anthropology Editing
- Law Paper Editing
- Engineering Paper Editing
- Technical Paper Editing
- Philosophy Editing
- PhD Dissertation Proofreading
- Lektorat Englisch
- Akademisches Lektorat
- Lektorat Englisch Preise
- Wissenschaftliches Lektorat
- Lektorat Doktorarbeit
PhD Thesis Editing
- Thesis Proofreading Services
- PhD Thesis Proofreading
- Proofreading Thesis Cost
- Proofreading Thesis
- Thesis Editing Services
- Professional Thesis Editing
- Thesis Editing Cost
- Proofreading Dissertation
- Dissertation Proofreading Cost
- Dissertation Proofreader
- Correção de Artigos Científicos
- Correção de Trabalhos Academicos
- Serviços de Correção de Inglês
- Correção de Dissertação
- Correção de Textos Precos
- 定額 ネイティブチェック
- Copy Editing
- FREE Courses
- Revision en Ingles
- Revision de Textos en Ingles
- Revision de Tesis
- Revision Medica en Ingles
- Revision de Tesis Precio
- Revisão de Artigos Científicos
- Revisão de Trabalhos Academicos
- Serviços de Revisão de Inglês
- Revisão de Dissertação
- Revisão de Textos Precos
- Corrección de Textos en Ingles
- Corrección de Tesis
- Corrección de Tesis Precio
- Corrección Medica en Ingles
- Corrector ingles
Select Page
Acknowledgements Example for an Academic Research Paper
Posted by Rene Tetzner | Sep 1, 2021 | How To Get Published | 0 |

Acknowledgements Example for an Academic or Scientific Research Paper This example of acknowledgements for a research paper is designed to demonstrate how intellectual, financial and other research contributions should be formally acknowledged in academic and scientific writing. As brief acknowledgements for a research paper, the example gathers contributions of different kinds – intellectual assistance, financial support, image credits etc. – into a single Acknowledgements section. Do note, however, that the formats preferred by some scholarly journals require the separation of certain contributions such as financial support of research into their own sections.

Although authors often write acknowledgements hastily, the Acknowledgements section is an important part of a research paper. Acknowledging assistance and contributions establishes your integrity as a researcher as well as your connections and collaborations. It can also help your readers with their own research, affect the influence and impact of the researchers and other professionals you thank, and demonstrate the value and purpose of the agencies that fund your work. The contents of the example I have prepared here are appropriate for a research paper intended for publication in a peer-reviewed journal, but the author, the research project, the manuscript studied, the journal publishing the paper and all those to whom gratitude is extended are entirely fictional. They were created for the purpose of demonstrating the following key concerns when writing the acknowledgements for a formal research paper:

• Writing in the first person (‘I’ for a single author or ‘we’ for two or more) to offer concise but sincere acknowledgements of specific contributions to your research. • Maintaining formal language, complete sentences and a professional tone to give specific and thorough information about contributions and convey collegial gratitude. • Expressing respect and appreciation in an appropriate fashion for each and every contribution and avoiding artificial or excessive flattery. • Using the complete names and preferred name formats for individuals, funding agencies, libraries, businesses and other organisations. Here, for example, I posit that the library holding the relevant manuscript has indicated that the name of the collection (lengthy though it is) should not be abbreviated. • Acknowledging contributions to your research and paper in the order that best represents the nature and importance of those contributions. The assistance of the author’s mentor comes first here, for instance, whereas the language editor is acknowledged much further down the list. • Meeting the requirements for acknowledgements set by the journal or other publisher of the research paper. For the example below, the goal is to record all relevant contributions to the research and paper in a single brief Acknowledgements section of 500 words or less – a set of parameters that would suit the acknowledgement requirements or expectations of many academic and scientific journals and even fit into a footnote or endnote if necessary.

Example Acknowledgements for an Academic Research Paper This paper and the research behind it would not have been possible without the exceptional support of my supervisor, Lawrence Magister. His enthusiasm, knowledge and exacting attention to detail have been an inspiration and kept my work on track from my first encounter with the log books of British Naval Ships MS VII.2.77 to the final draft of this paper. Margaret Kempis and Matthew Brown, my colleagues at Western University, have also looked over my transcriptions and answered with unfailing patience numerous questions about the language and hands of British Naval Ships MS VII.2.77. Samantha McKenzie, head librarian of the Southern Region Central Collegiate Library Special Collections and Microfilms Department where British Naval Ships MS VII.2.77 currently resides, not only provided colour images of the manuscript overnight, but unexpectedly shared the invaluable information on the book that she has been gathering for almost twenty years. I am also grateful for the insightful comments offered by the anonymous peer reviewers at Books & Texts. The generosity and expertise of one and all have improved this study in innumerable ways and saved me from many errors; those that inevitably remain are entirely my own responsibility.
Studying British Naval Ships MS VII.2.77 has proved extremely costly and I am most thankful for the Western University Doctoral Fellowship that has provided financial support for the larger project from which this paper grew. A travel grant from the Literary Society of the Southern Region turned the hope of working in person with British Naval Ships MS VII.2.77 into a reality, and the generous offer of free accommodation from Ms McKay (Samantha McKenzie’s aunt) allowed me to continue my research with the book much longer than I could have hoped. The final design of the complicated transcription tables in Appendices I–III is the creative and technical work of Sam Stone at A+AcaSciTables.com, and the language and format of the paper have benefited enormously from the academic editing services of Veronica Perfect. Finally, it is with true pleasure that I acknowledge the contributions of my amazing partner, Kendric James, who has given up many a Friday evening and Sunday afternoon to read every version of this paper and the responses it has generated with a combination of compassion and criticism that only he could muster for what he fondly calls ‘my odd obsession with books about the sea.’
You might be interested in Services offered by Proof-Reading-Service.com
Journal editing.
Journal article editing services
PhD thesis editing services
Scientific Editing
Manuscript editing.
Manuscript editing services
Expert Editing
Expert editing for all papers
Research Editing
Research paper editing services
Professional book editing services
Related Posts

Choosing the Right Journal
September 10, 2021

Example of a Quantitative Research Paper
September 4, 2021

What Is a Good H-Index Required for an Academic Position?
September 3, 2021

Free Sample Letters for Withdrawing a Manuscript
August 31, 2021
Our Recent Posts

Our review ratings
- Examples of Research Paper Topics in Different Study Areas Score: 98%
- Dealing with Language Problems – Journal Editor’s Feedback Score: 95%
- Making Good Use of a Professional Proofreader Score: 92%
- How To Format Your Journal Paper Using Published Articles Score: 95%
- Journal Rejection as Inspiration for a New Perspective Score: 95%
Explore our Categories
- Abbreviation in Academic Writing (4)
- Career Advice for Academics (5)
- Dealing with Paper Rejection (11)
- Grammar in Academic Writing (5)
- Help with Peer Review (7)
- How To Get Published (146)
- Paper Writing Advice (17)
- Referencing & Bibliographies (16)
Recognizing Contributions: Acknowledge In Research Paper
Learn how to acknowledge in research paper with our simple guide, and ensure that your work gains recognition.
When we work on any project, it’s not just about our individual effort, it’s about teamwork as well. It’s important to acknowledge the contributions of others who have helped you along the way. This is where the acknowledgment section comes in. In this part of your paper, you can express your gratitude to those who have supported you throughout the research process, such as funders, advisors, assistants, collaborators, participants, and editors.
However, it’s important to be mindful of ethical considerations and avoid any promotion or advertising of specific individuals or organizations. In this guide, we’ll cover everything you need to know about acknowledgments, including how to acknowledge in research paper , tips for writing them, common mistakes to avoid, and ethical considerations. So, let’s get started!
What Is Acknowledgement In A Research Paper?
The acknowledgment section in a research paper credits individuals, institutions, or organizations that aided in the research or manuscript preparation. It’s usually found after the conclusion.
While optional, acknowledgments are commonly added to recognize and thank contributors for their efforts. This section typically starts with a statement thanking those who funded or supported the project, along with colleagues, research assistants, or other contributors who provided valuable feedback or assistance.
Being specific and detailing the contributions of each individual or organization is crucial to show their importance in the research process. However, it’s unnecessary to acknowledge everyone who helped, and acknowledgments should be kept brief and relevant to the project.
Why Is Acknowledgment Important?
Acknowledgment holds significant importance in research as it acknowledges and provides credit to individuals or organizations who have contributed to the research project. It expresses gratitude for their guidance, support, and assistance during the research process.
Acknowledgments are commonly included in research papers to build relationships and encourage future collaborations with those who have supported the research. By acknowledging their contributions, researchers demonstrate their appreciation for the input of others and the importance of collaboration in the research process.
Moreover, the acknowledgment section ensures academic integrity by recognizing and crediting all contributors to the research project. It also prevents any potential issues related to plagiarism or lack of attribution.
Who Should Be Acknowledged?
Acknowledgments in a research paper should recognize and give credit to individuals, organizations, or institutions that contributed to the research project in some way. This can include:
- Funding sources: Acknowledge those who provided financial support for the research project.
- Academic advisors or mentors: Acknowledge those who provided guidance or supervision throughout the research process.
- Research assistants: Acknowledge those who provided technical or administrative support during the research.
- Participants : Acknowledge those who took part in the research study, such as survey respondents.
- Collaborators : Acknowledge colleagues or other researchers who contributed to the research project in some way.
- Editors or proofreaders: Acknowledge those who helped with editing or proofreading the manuscript .
- Institutions or organizations: Acknowledge the institution or organization that provided resources or support for the research project.
Types Of Acknowledgement
Acknowledgment in a research paper can take various forms, depending on the purpose and context of the project. Here are some common types of acknowledgments:
1. Formal Acknowledgments
These are typically written in a formal tone and are used to recognize and give credit to people, organizations, or institutions that provide financial or technical support to the research project. These acknowledgments often appear at the beginning or end of the research paper and may include formal language and formatting.
2. Informal Acknowledgments
These acknowledgments are often more personal and informal in tone. They may include acknowledging friends, family members, or colleagues who provided emotional support or helped in some way during the research process.
3. Professional Acknowledgments
These acknowledgments are typically used in academic or professional settings and are aimed at giving credit to individuals or organizations that contributed to the research project. These acknowledgments may include thanking mentors, colleagues, research assistants, or funding agencies.
4. Collaborative Acknowledgments
These acknowledgments are used to recognize the collaborative nature of research projects. They may include acknowledging co-authors, collaborators, or other researchers who contributed to the project in some way.
Tips For Writing An Acknowledgement
When writing acknowledgment in a research paper, it’s important to keep the following tips in mind:
- Be specific: Clearly mention the contributions made by individuals or organizations, and how they helped in the research process.
- Use appropriate tone: Write in a professional tone and avoid using overly emotional language.
- Keep it concise: Avoid lengthy paragraphs and keep the acknowledgment section brief and relevant.
- Follow the required format: Check the guidelines provided by the journal or institution and ensure that you follow the required format.
- Proofread: Carefully proofread the acknowledgment section for any errors or typos.
- Be grateful: Show appreciation and gratitude to the individuals or organizations who contributed to the research project.
- Avoid self-promotion: The acknowledgment section should not be used to promote oneself or one’s organization.
Examples Of Acknowledgement
Examples of acknowledgments in a research paper include thanking the funding sources, academic advisors or mentors, research assistants, participants, collaborators, editors or proofreaders, and institutions or organizations that provided support. Here are some sample acknowledgments that are concise and relevant to the research project:
Acknowledge In Research Paper: Example 1
“I would like to thank Dr. Ram for his invaluable guidance and support throughout this project. I am also grateful to my research assistant, Priya, for her technical expertise and administrative assistance. This project would not have been possible without the generous financial support of the XYZ Foundation. Lastly, I would like to acknowledge the study participants who generously shared their time and insights.”
Acknowledge In Research Paper: Example 2
“I am indebted to Dr. Mary for her continuous support and feedback throughout the research process. I also want to thank my colleagues, Shahin and Sarah, for their valuable input and suggestions. The editorial assistance provided by XYZ Editing Services was also greatly appreciated. I am also grateful to the ABC Institution for providing the necessary resources for this research project.”
Common Mistakes To Avoid When Writing An Acknowledgement
When writing acknowledgment, it’s important to avoid certain mistakes, such as:
- Forgetting to acknowledge someone who contributed to the research project.
- Using vague language instead of specific details about how someone contributed.
- Focusing too much on personal anecdotes or stories, rather than keeping the acknowledgment concise and relevant to the research project.
- Using the acknowledgment section to promote or advertise specific individuals or organizations.
- Forgetting to proofread the acknowledgment section for errors in grammar or spelling.
- Including acknowledgments that are not relevant to the research project.
- Making it too formal or too casual, rather than matching the tone of the rest of the research paper.
To avoid these mistakes, it’s important to carefully consider who should be acknowledged, what specific contributions they made, and to keep the language concise and relevant to the research project. It’s also helpful to have someone else review the acknowledgment section to ensure that it’s free of errors and strikes the right tone.
How To Acknowledge In Research Paper?
- When citing an acknowledgment in a research paper, it should be listed as a separate section at the end of the paper, following the references section. It should be titled “Acknowledgement” and be placed after the conclusion but before the reference list.
- Acknowledgment section should not be included within the text citation or reference list. However, if a person or organization mentioned in the acknowledgment section was cited within the text, it should be included in the in-text citation and reference list.
- It’s important to make sure that acknowledgments are cited correctly in order to give credit to those who contributed to the research project. This will help to ensure academic integrity and avoid plagiarism. Learn more about Plagiarism here .
Ethical Considerations For Acknowledging Others In Your Research Paper
It is essential to consider ethical principles when acknowledging others in your research paper. First and foremost, ensure that you acknowledge all individuals and organizations that made significant contributions to your research. This acknowledgment must be honest and accurate and should not falsely claim credit for the work of others.
Additionally, it is crucial to obtain consent from individuals before acknowledging them in your research paper, particularly when using their personal information. Ensure that you have informed them about how their contribution will be acknowledged and seek their permission to do so.
The Best Infographic Maker And Overall Full-Stack Design Tool
Have you ever wondered how confused your audience would be if you published a plain text-based research paper with complex terms used within it? This is where the importance of visuals and graphics comes in. They help amplify your work by breaking the information with pictorial representation. Mind the Graph is the best infographic maker and full-stack design tool that helps to exemplify your research work. Sign up for free now!
Subscribe to our newsletter
Exclusive high quality content about effective visual communication in science.
About Sowjanya Pedada
Sowjanya is a passionate writer and an avid reader. She holds MBA in Agribusiness Management and now is working as a content writer. She loves to play with words and hopes to make a difference in the world through her writings. Apart from writing, she is interested in reading fiction novels and doing craftwork. She also loves to travel and explore different cuisines and spend time with her family and friends.
Content tags
Writing Acknowledgments for Your Research Paper
- Research Process
- Peer Review
In this article, we describe what types of contributions warrant mention in the acknowledgments section of a paper .
Updated on July 8, 2014

In another article , we discuss four criteria that must be met for an individual to qualify for manuscript authorship. In this article, we describe what types of contributions warrant mention in the acknowledgments section of a paper instead. The International Committee of Medical Journal Editors (ICMJE) describes several roles that merit acknowledgment, rather than authorship :
“acquisition of funding; general supervision of a research group or general administrative support; and writing assistance, technical editing, language editing, and proofreading.”
You should also acknowledge direct technical assistance, including help with animals, cells, equipment, patients, procedures, or techniques or provision of data, equipment, reagents, or samples, as well as more indirect assistance via intellectual discussions. Note that all of these contributions are typically more mechanical, indirect, and/or one-dimensional than those of authors. Additionally, some argue that individuals who provided help and could be chosen as a peer reviewer, leading to a potential conflict of interest, should be cited.
In any case, the ICMJE states that contributors may be cited individually or collectively and that their precise contributions should be specified.
e.g., “We thank Dr. X and Dr. Y for performing the surgeries” or “We thank the physicians who performed the surgeries"
Institutional affiliations may or may not be mentioned, depending on the journal's guidelines. Finally, the ICMJE encourages written permission from acknowledged individuals “because acknowledgment may imply endorsement.”
Funding sources should also be mentioned in the acknowledgments section, unless your target journal requires a separate section for this information. Whether the funding was partial or full, relevant grant numbers, and the author(s) who received the funding, if applicable, should be detailed as well. Note that acknowledging grants and fellowships is in fact required by many funding agencies and research institutions.
In contrast, contributions that are not specifically related to your research, including personal encouragement (e.g., by your friends or parents) and very general help (e.g., from a laboratory manager who purchases all supplies for your research group), should not be cited. Additionally, anonymous editors and peer reviewers are usually not thanked in the acknowledgments section; many journals (such as American Physical Society journals ) explicitly discourage this practice because it is difficult to comprehensively acknowledge all anonymous support and because this practice could potentially bias reviewers.
The writing style of acknowledgments sections may vary according to the journal, but generally, these sections are written in the first person and are as succinct as possible. A statement about conflicts of interest, citation of previous publication in poster or abstract form, and other information may also be included in this section, again depending on the journal. As you proceed through revisions for one journal or if you change your target journal, remember to reformat as necessary and to update your acknowledgments if additional help was obtained during the revision, such as with editing or new experiments.
Although an acknowledgments section may be appended to the end of your manuscript or relegated to a footnote, it is not a trivial component. By acknowledging all help received with your research, you are demonstrating your integrity as a researcher, which in turn encourages continued collaboration. You may also be bolstering your colleagues' careers, as being credited in an acknowledgments section is emerging as one of many gauges of a researcher's professional impact beyond citations (see ImpactStory , based on altmetrics ). Furthermore, information about who provided certain data, equipment, protocols, reagents, or samples may be of help to other researchers in your field.
This editing tip has hopefully elucidated what to include in the acknowledgments section of your manuscript and why this section is significant. If you have any comments or questions, please contact us . Best wishes in your research and writing!

Michaela Panter, PhD
See our "Privacy Policy"

Acknowledgement in Research Paper | How to Write | Perfect Example
What is acknowledgement in research paper.
Acknowledgement in a research paper is the section where the author expresses gratitude to individuals and organizations who have contributed to the completion of the study. This section is usually placed at the beginning or end of the paper and is an important part of the research process. It allows the author to recognize the support, assistance, and guidance they have received from others in the course of their research.
What is the purpose of acknowledgement in research paper?
The acknowledgement section is an opportunity for the author to show appreciation for anyone who has helped them in the research process, including mentors, advisors, colleagues, and funding agencies. It is also a way to acknowledge the contributions of participants, interviewees, or anyone else who has played a role in the study.
Acknowledgements can also include thanks to individuals who have provided critical feedback, technical assistance, or resources that have been essential to the research project. Overall, this section is a way for the author to show their appreciation for the collaborative and supportive nature of the research community.
How to write acknowledgement in research paper?
- Identify Key Contributors : Make a list of individuals and organizations that have contributed significantly to your research. This includes advisors, mentors, collaborators, funders, participants, and institutions.
- Understand the Purpose : Acknowledgments are meant to recognize and thank those who have supported or contributed to your research in various ways. Understand the purpose of this section is to express gratitude and recognize their contributions.
- Be Genuine and Specific : Your acknowledgments should be sincere and specific. Avoid generic expressions of thanks and instead, mention the specific contributions each person or organization made to your research.
- Start with Formality : Begin your acknowledgment section with a formal tone and expression of gratitude. Address individuals with their appropriate titles, such as Dr., Prof., Mr., or Ms., and mention any institutional affiliations if relevant.
- Personalize Your Thanks : Tailor your acknowledgments to acknowledge each individual’s specific contributions. Mention how their support, guidance, or expertise influenced your research or contributed to its success.
- Maintain Professionalism : While acknowledgments can be personal, maintain a professional tone and avoid overly informal language or colloquialisms. Remember that this section is part of your scholarly work.
- Consider Cultural Sensitivities : Be mindful of cultural norms and sensitivities when expressing gratitude, especially if your research involves international collaborations. Tailor your acknowledgments to reflect cultural expectations or customs, as appropriate.
- Proofread Carefully : Ensure your acknowledgments are free of grammatical errors and typos. Take the time to review and edit this section to ensure clarity and coherence.
- Respect Space Limitations : While it’s important to acknowledge all key contributors, be mindful of space limitations in your research paper. Prioritize mentioning those who made significant contributions while being respectful of length constraints.
- End on a Positive Note : Conclude your acknowledgment section with a positive and appreciative tone. Express your gratitude to everyone who supported your research journey, including family, friends, colleagues, and institutions.
Example of Acknowledgement in Research
I am immensely grateful to the individuals and organizations whose support and guidance have been instrumental in the completion of this research paper. Their assistance and expertise have significantly contributed to the development and refinement of my study.
Firstly, I extend my heartfelt appreciation to Dr. Aurora Rivera, my mentor and advisor throughout this research journey. Her insightful feedback, encouragement, and scholarly guidance have been invaluable in shaping the direction and methodology of my study.
I am also deeply thankful to Mr. Santiago Cruz, Director of Research at the Philippine Social Sciences Institute, for his generous support and assistance in accessing research resources. His expertise and encouragement have greatly facilitated the progress of my research.
Special gratitude is extended to my research collaborators, including Dr. Sofia Ramirez and Mr. Miguel Hernandez, for their collaborative efforts and insightful perspectives that have enriched the depth and analysis of my research findings.
I would like to acknowledge the valuable contributions of the staff and researchers at the Philippine Research Center for Social Sciences, whose support and expertise have been indispensable in navigating complex research methodologies and data analysis.
Furthermore, I extend my sincere appreciation to the participants of my study for their cooperation and willingness to share their experiences, which have provided essential insights and perspectives.
I wish to express my profound gratitude to my family and friends for their unwavering support, encouragement, and understanding throughout this research endeavor. Their belief in my abilities has been a constant source of motivation and inspiration.

Acknowledgement in Research Paper Example
We extend our heartfelt gratitude to the individuals and institutions whose unwavering support has been indispensable in the completion of this research paper. Their guidance, encouragement, and expertise have greatly contributed to the success of our study.
Firstly, we express our sincere appreciation to Dr. Juanita Cruz, Professor of Economics at the University of the Philippines, for her invaluable mentorship and insightful feedback throughout the research process. Her expertise in the field has been instrumental in shaping the direction of our study.
We are also indebted to Dr. Manuel Reyes, Director of the Philippine Economic Development Institute, for his generous assistance and provision of research resources. His support has been pivotal in enhancing the quality and depth of our analysis.
Special thanks are extended to our research collaborators, including Dr. Sofia Garcia from the Department of Political Science at Ateneo de Manila University, and Dr. Miguel Hernandez from the Department of Sociology at Mapua University. Their collaboration and expertise have enriched our research with diverse perspectives and profound insights.
We would like to acknowledge the invaluable assistance provided by the staff and researchers at the Philippine Institute for Development Studies, who generously shared their knowledge and resources throughout the duration of our study.
Furthermore, we extend our gratitude to the participants of our study, whose cooperation and willingness to share their experiences have been instrumental in shaping our findings.
- Acknowledgement for Thesis
- Acknowledgement for Dissertation
Similar Posts
Acknowledgement for portfolio | how to write | with sample.
Acknowledgement for portfolio is crucial for recognizing the contributions and efforts of others. It serves as a way…
Acknowledgement for Investigatory Project [5 Samples with PDF]
Acknowledgement in an investigatory project play a crucial role in recognizing and appreciating the individuals or organizations that…
Acknowledgement for Internship Report 📝
Writing an acknowledgment for your internship report may seem like a small and insignificant task, but it actually…
Acknowledgement for Thesis [Sample and Best Practice]
What is Acknowledgement in Thesis? Writing a thesis is a significant milestone in a student’s academic journey. A…
Acknowledgement for Assignment (6 Sampels with PDF)
Acknowledgement is a pivotal aspect of any assignment, showcasing gratitude towards those who have contributed to its completion….
Acknowledgement for Dissertation [With Example]
Writing a dissertation is a significant academic undertaking that requires extensive research, critical analysis, and original contributions to…
.webp)
How to Write an Acknowledgement for a Research Paper

Hey guys, Phill Collins here! Today, I will teach you how to write an acknowledgment section in a research paper. Let’s do this!
Acknowledging contributions is a crucial aspect of creating a thorough research paper. It provides an opportunity to convey appreciation and acknowledge the support from individuals and institutions throughout your work. This piece will explore the intricacies of crafting acknowledgments for papers, offering valuable insights, practical advice, and sample acknowledgments. It aims to assist you in expressing gratitude to those who have played a substantial role in your research journey. As usual, I recommend those of you who struggle with your writings to pay for a research paper to save time and have a stress-free evening.
What Is Acknowledgement in a Research Paper
Acknowledgment in a research paper is a section dedicated to expressing gratitude and recognizing the individuals, institutions, or resources that have contributed to the completion of the research. This section is an opportunity for the author to appreciate the support, guidance, or assistance received during the research process. Acknowledgments go beyond the academic content of the paper and serve as a personal and professional gesture of recognition for those who played a significant role in the research endeavor.
In this section, researchers typically acknowledge the contributions of mentors, advisors, colleagues, or peers who provided valuable insights, feedback, or assistance in shaping the research project. Additionally, institutions, funding agencies, or organizations that supported the research financially or through resources may be acknowledged. The acknowledgment section reflects the collaborative and communal nature of academic work, highlighting the interconnected web of individuals and entities that contribute to the scholarly pursuit.
While there is no strict format for writing acknowledgments, it is important to balance professionalism and sincerity. Authors can use this space to express genuine gratitude, share personal reflections on the collaborative process, and convey the impact of the support received. The acknowledgment section adds a human touch to the paper, recognizing the collective effort that goes into the creation of academic knowledge.

The Role of an Acknowledgment in a Research Papers
The acknowledgment section in a paper plays a vital role in recognizing and appreciating the various contributors and influences that have shaped the research journey. Beyond the academic rigor captured in the main body of the paper, acknowledgments offer a space to express gratitude for the support and guidance received during the research process. This section often serves as a heartfelt acknowledgment of the collaborative effort to bring a research project to fruition.
Essentials of an Acknowledgement in Research Paper
An essential component of a complex paper, the acknowledgment section serves as a heartfelt expression of gratitude towards individuals and entities who have contributed significantly to the research process. In this section, authors typically recognize mentors, advisors, colleagues, and peers who provided valuable insights, guidance, or support. Additionally, institutions, funding sources, or organizations that played a role in the research project are acknowledged. The acknowledgment is a personal touch within the scholarly document, acknowledging the collaborative nature of academic work and underscoring the importance of communal support in the research journey. It adds a human element to the paper, recognizing the interconnected network of individuals and resources that contribute to the scholarly endeavor.
How to Write an Acknowledgement for Research Paper Using 8 Simple Tips
Keep in mind that acknowledgments offer a chance to express gratitude and acknowledge the valuable contributions of those who assisted you. Dedicate time to creating a genuine and thoughtful acknowledgment section that mirrors the collaborative and supportive nature of your research endeavor.
Sincerity and Authenticity
Write your acknowledgments with heartfelt gratitude, conveying genuine appreciation for the support and contributions you received throughout your research journey.

Specific Individuals and Institutions
Identify key figures and entities that played a substantial role in your research, including mentors, advisors, collaborators, and funding agencies. Acknowledge institutions that provided resources or facilities.
Conciseness and Focus
Keep your acknowledgments brief and to the point. Concentrate on highlighting the pivotal individuals and organizations that significantly influenced your research.
Formal Tone
Maintain a professional and formal tone throughout your acknowledgments. Remember that this section serves as a formal recognition of support, not a casual conversation.
Ethical Considerations
Adhere to ethical guidelines and norms when acknowledging individuals and organizations. Respect the privacy and confidentiality of those who may prefer not to be mentioned.
Personal Touches
If appropriate, include personal anecdotes or specific instances where individuals or organizations made a noteworthy impact on your research.
Tailor to Research Context
Consider the nature of your research and customize your acknowledgments accordingly. For instance, if your research is interdisciplinary, recognize experts from various fields who provide valuable insights.
Appreciation Beyond Formal Requirements
While an acknowledgement in research paper has to list individuals and institutions, extend your appreciation to others who indirectly supported you. This may include family, friends, or colleagues who provided emotional support during your research journey.
Example of Acknowledgement in Research Paper
.webp)
An Additional Example of Acknowledgement in Research Paper
.webp)
Final Words
In my opinion, acknowledgements in a research paper provide an avenue to convey appreciation and recognize the indispensable contributions of individuals and institutions that have bolstered your work. In this article, I did my best to offer tips and a sample acknowledgment to assist you in authentically expressing your gratitude. Keep in mind that acknowledgments are a chance to genuinely convey appreciation and attribute credit where it is rightfully due.
How to write acknowledgement in research paper?
Begin your acknowledgment section with a formal salutation, expressing gratitude to those who contributed to your research. Use a sincere and appreciative tone, mentioning specific individuals and institutions, and keep it concise.
What is the purpose of the acknowledgement section in a research paper?
The acknowledgment section serves to express gratitude and recognize the contributions of individuals and institutions who supported the research. It reflects the researcher's appreciation for guidance, resources, and collaboration during the project.
What should the acknowledgement section of a research paper include?
An acknowledgement for research paper includes thanks to advisors, committee members, funding agencies, collaborators, institutions providing resources, and anyone who significantly contributed. Maintain a formal tone, adhere to ethical considerations, and, if appropriate, add a personal touch or anecdotes.
How to acknowledge someone in a research paper?
Acknowledge individuals by mentioning their names, roles, and specific contributions. Follow a formal and respectful tone, adhering to ethical guidelines. If applicable, express personal appreciation and consider tailoring acknowledgments to the nature of the research and relationships involved.
Frequently asked questions
She was flawless! first time using a website like this, I've ordered article review and i totally adored it! grammar punctuation, content - everything was on point
This writer is my go to, because whenever I need someone who I can trust my task to - I hire Joy. She wrote almost every paper for me for the last 2 years
Term paper done up to a highest standard, no revisions, perfect communication. 10s across the board!!!!!!!
I send him instructions and that's it. my paper was done 10 hours later, no stupid questions, he nailed it.
Sometimes I wonder if Michael is secretly a professor because he literally knows everything. HE DID SO WELL THAT MY PROF SHOWED MY PAPER AS AN EXAMPLE. unbelievable, many thanks
You Might Also Like
.png)
New Posts to Your Inbox!
Stay in touch
Have a language expert improve your writing
Run a free plagiarism check in 10 minutes, automatically generate references for free.
- Knowledge Base
- Dissertation
- Thesis & Dissertation Acknowledgements | Tips & Examples
Thesis & Dissertation Acknowledgements | Tips & Examples
Published on 4 May 2022 by Tegan George . Revised on 4 November 2022.
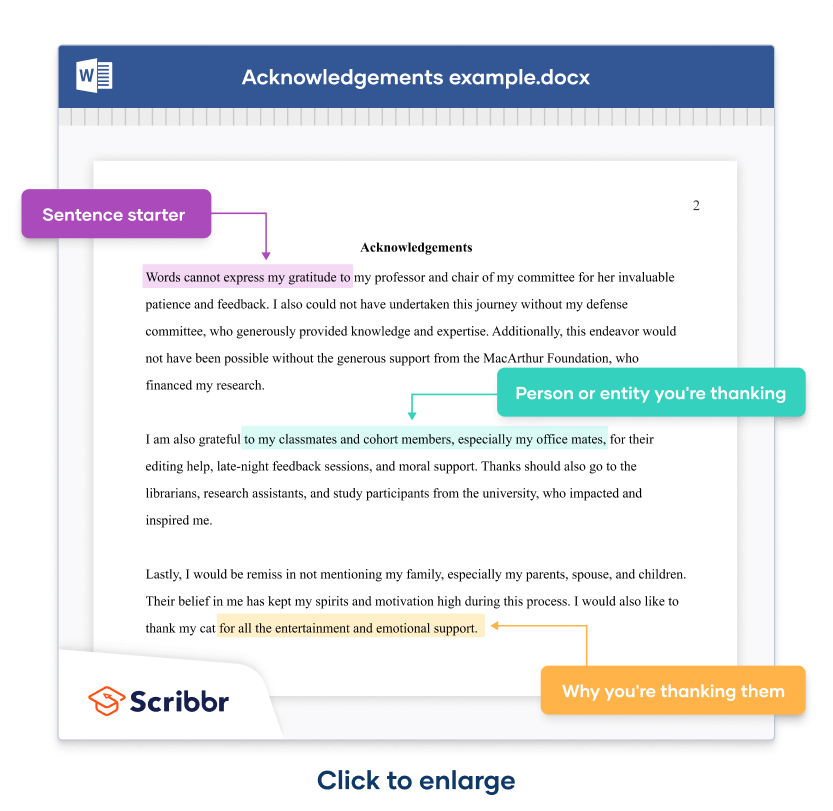
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process.
Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page.
In your acknowledgements, it’s okay to use a more informal style than is usually permitted in academic writing , as well as first-person pronouns . Acknowledgements are not considered part of the academic work itself, but rather your chance to write something more personal.
To get started, download our step-by-step template in the format of your choice below. We’ve also included sample sentence starters to help you construct your acknowledgments section from scratch.
Download Word doc Download Google doc
Instantly correct all language mistakes in your text
Be assured that you'll submit flawless writing. Upload your document to correct all your mistakes.

Table of contents
Who to thank in your acknowledgements, how to write acknowledgements, acknowledgements section example, acknowledgements dos and don’ts, frequently asked questions.
Generally, there are two main categories of acknowledgements: professional and personal .
A good first step is to check your university’s guidelines, as they may have rules or preferences about the order, phrasing, or layout of acknowledgements. Some institutions prefer that you keep your acknowledgements strictly professional.
Regardless, it’s usually a good idea to place professional acknowledgements first, followed by any personal ones. You can then proceed by ranking who you’d like to thank from most formal to least.
- Chairs, supervisors, or defence committees
- Funding bodies
- Other academics (e.g., colleagues or cohort members)
- Editors or proofreaders
- Librarians, research/laboratory assistants, or study participants
- Family, friends, or pets
Typically, it’s only necessary to mention people who directly supported you during your thesis or dissertation. However, if you feel that someone like a secondary school physics teacher was a great inspiration on the path to your current research, feel free to include them as well.
Professional acknowledgements
It is crucial to avoid overlooking anyone who helped you professionally as you completed your thesis or dissertation. As a rule of thumb, anyone who directly contributed to your research should be mentioned.
A few things to keep in mind include:
- Even if you feel your chair didn’t help you very much, you should still thank them first to avoid looking like you’re snubbing them.
- Be sure to follow academic conventions, using full names with titles where appropriate.
- If several members of a group or organisation assisted you, mention the collective name only.
- Remember the ethical considerations around anonymised data. If you wish to protect someone’s privacy, use only their first name or a generic identifier (such as ‘the interviewees’).
Personal acknowledgements
There is no need to mention every member of your family or friend group. However, if someone was particularly inspiring or supportive, you may wish to mention them specifically. Many people choose to thank parents, partners, children, friends, and even pets, but you can mention anyone who offered moral support or encouragement, or helped you in a tangible or intangible way.
Some students may wish to dedicate their dissertation to a deceased influential person in their personal life. In this case, it’s okay to mention them first, before any professional acknowledgements.
The only proofreading tool specialized in correcting academic writing
The academic proofreading tool has been trained on 1000s of academic texts and by native English editors. Making it the most accurate and reliable proofreading tool for students.
Correct my document today
After you’ve compiled a list of who you’d like to thank, you can then sort your list into rank order. Separate everyone you listed into ‘major thanks’, ‘big thanks’, and ‘minor thanks’ categories.
- ‘Major thanks’ are given to people who your project would be impossible without. These are often predominantly professional acknowledgements, such as your advisor , chair, and committee, as well as any funders.
- ‘Big thanks’ are an in-between, for those who helped you along the way or helped you grow intellectually, such as classmates, peers, or librarians.
- ‘Minor thanks’ can be a catch-all for everyone else, especially those who offered moral support or encouragement. This can include personal acknowledgements, such as parents, partners, children, friends, or even pets.
How to phrase your acknowledgements
To avoid acknowledgements that sound repetitive or dull, consider changing up your phrasing. Here are some examples of common sentence starters you can use for each category.
Note that you do not need to write any sort of conclusion or summary at the end. You can simply end the acknowledgements with your last thank-you.
Here’s an example of how you can combine the different sentences to write your acknowledgements.
A simple construction consists of a sentence starter (in purple highlight ), followed by the person or entity mentioned (in green highlight ), followed by what you’re thanking them for (in yellow highlight .)
Acknowledgements
Words cannot express my gratitude to my professor and chair of my committee for her invaluable patience and feedback. I also could not have undertaken this journey without my defense committee, who generously provided knowledge and expertise. Additionally, this endeavor would not have been possible without the generous support from the MacArthur Foundation, who financed my research .
I am also grateful to my classmates and cohort members, especially my office mates, for their editing help, late-night feedback sessions, and moral support. Thanks should also go to the librarians, research assistants, and study participants from the university, who impacted and inspired me.
Lastly, I would be remiss in not mentioning my family, especially my parents, spouse, and children. Their belief in me has kept my spirits and motivation high during this process. I would also like to thank my cat for all the entertainment and emotional support.
- Write in first-person, professional language
- Thank your professional contacts first
- Include full names, titles, and roles of professional acknowledgements
- Include personal or intangible supporters, like friends, family, or even pets
- Mention funding bodies and what they funded
- Appropriately anonymise or group research participants or non-individual acknowledgments
Don’t:
- Use informal language or slang
- Go over one page in length
- Mention people who had only a peripheral or minor impact on your work
You may acknowledge God in your thesis or dissertation acknowledgements , but be sure to follow academic convention by also thanking the relevant members of academia, as well as family, colleagues, and friends who helped you.
Yes, it’s important to thank your supervisor(s) in the acknowledgements section of your thesis or dissertation .
Even if you feel your supervisor did not contribute greatly to the final product, you still should acknowledge them, if only for a very brief thank you. If you do not include your supervisor, it may be seen as a snub.
In the acknowledgements of your thesis or dissertation, you should first thank those who helped you academically or professionally, such as your supervisor, funders, and other academics.
Then you can include personal thanks to friends, family members, or anyone else who supported you during the process.
The acknowledgements are generally included at the very beginning of your thesis or dissertation, directly after the title page and before the abstract .
In a thesis or dissertation, the acknowledgements should usually be no longer than one page. There is no minimum length.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the ‘Cite this Scribbr article’ button to automatically add the citation to our free Reference Generator.
George, T. (2022, November 04). Thesis & Dissertation Acknowledgements | Tips & Examples. Scribbr. Retrieved 9 April 2024, from https://www.scribbr.co.uk/thesis-dissertation/acknowledgements/
Is this article helpful?
Tegan George
Other students also liked, dissertation title page, how to write an abstract | steps & examples, dissertation table of contents in word | instructions & examples.
We use cookies on this site to enhance your experience
By clicking any link on this page you are giving your consent for us to set cookies.
A link to reset your password has been sent to your email.
Back to login
We need additional information from you. Please complete your profile first before placing your order.
Thank you. payment completed., you will receive an email from us to confirm your registration, please click the link in the email to activate your account., there was error during payment, orcid profile found in public registry, download history, what to include in your acknowledgments section.
- Charlesworth Author Services
- 02 June, 2018
- Academic Writing Skills
Most academic papers have many people who have helped in some way in the preparation of the written version or the research itself. This could be someone from a sponsoring institution, a funding body, other researchers, or even family, friends or colleagues who have helped in the preparation. These people need to be mentioned in the Acknowledgments section of the paper.
Acknowledgments section in different academic documents
The Acknowledgments section is present in both a paper and an academic thesis . For papers, the Acknowledgments section is usually presented at the back, whereas in a thesis, this section is located towards the front of the manuscript and is commonly placed somewhere between the abstract and Introduction . However, the exact location varies between each university , as each establishment possesses its own style guide for theses and student submissions. So, it is always worthwhile consulting your university’s academic style guide before writing a manuscript for undergraduate/postgraduate submission.
Acknowledgments section in theses
For academic theses, there is no right or wrong way to acknowledge people, and who you want to acknowledge is down to personal preference. However, the common types of people authors acknowledge in their academic theses include:
- Supervisor’s contributions
- Research group (especially if the thesis in question is a master’s and the work is helped along by a PhD student)
- Support staff (laboratory technicians, etc.)
- Any students who undertook side projects with them (e.g. final year undergraduates, summer students, master’s students)
- Administrative staff (there can be a lot of bureaucracy for thesis submissions)
- Referees that got them onto the course (postgraduate only)
- Funding bodies
- Any collaboration with industry and the people they worked with at said establishment(s)
Acknowledgments section in journal papers
Now, whilst university manuscripts can include any combination of the above (including all and none in some cases), academic publications in journals more commonly acknowledge the same kind of people/organizations, but again it is up to the author(s) what they feel should be acknowledged; not every piece of help needs to be acknowledged, just the most useful/prevalent help. Also, acknowledgments should be written in the first person .
Examples of whom and what should be acknowledged in a journal publication are listed below:
- Direct technical help (e.g. supply of animal subjects, cells, equipment setup, methods , statistics/data manipulation, samples, chemicals/reagents, analytical/spectroscopy techniques)
- Indirect assistance (topical and intellectual discussions about the research which can lead to generation of new ideas)
- Affiliated institutions
- Grant numbers
- Who received the funding (if not the author, e.g. a supervisor)
- Any associated fellowships
Whom to acknowledge - and whom not to acknowledge
- Other authors/contributors : It is not common practice for the lead paper writer (i.e. the person writing and publishing the manuscript) to acknowledge the other authors/direct contributors to the paper. Only those who are not recognized as authors may be thanked and acknowledged.
- Reviewers : Authors are also not allowed to thank reviewers personally, or those who inspire them but cannot directly receive their appreciation – although reviewers can be thanked if they are kept anonymous .
- Friends and family : Unlike university manuscripts, journal manuscripts should not include help and guidance from family and friends.
Other acknowledgments
- Titles and institutions : Titles such as Mr, Mrs, Miss, etc. are not commonly included, but honorary titles such as Dr, Professor, etc. are. The institutions of the acknowledged people are usually mentioned.
For example, the following would not be acceptable:
We dedicate this work to the deceased Prof. Bloggs.
However, the following would be acceptable:
We acknowledge Prof. Bloggs for discovering the secret of anonymity.
Additional pointers for writing the Acknowledgments section
- The tone of the section should be in an active voice.
- Do not use pronouns indicating possession (i.e. his, her, their, etc.).
- Terms associated with specific companies should be written out in full, e.g. Limited, Corporation, etc.
- If the results have been published elsewhere, then this should also be acknowledged.
- Any abbreviations should be expanded unless the abbreviation appears in the main body of the text.
Below are examples of the Acknowledgments sections taken from a couple of papers from Nature Communications :

Duan L., Hope J., Ong Q., Lou H-Y., Kim N., McCarthy C., Acero V., Lin M., Cui B., Understanding CRY2 interactions for optical control of intracellular signalling, Nature Communications, 2017, 8:547
Xu Q., Jensen K., Boltyanskiy R., Safarti R., Style R., Dufresne E., Direct measurement of strain-dependent solid surface stress, Nature Communications, 2017, 8:555
Many people think that the Acknowledgments section of a manuscript is a trivial and unimportant component. However, it constitutes a vital means to ensure that all affiliated support for the paper can be duly and transparently mentioned. By acknowledging people for their efforts and contributions, you demonstrate your integrity as an academic researcher. In addition, crediting other people for their help can also increase their presence in the academic world and possibly help to boost their career as well as your own.
Maximise your publication success with Charlesworth Author Services.
Charlesworth Author Services, a trusted brand supporting the world’s leading academic publishers, institutions and authors since 1928.
To know more about our services, visit: Our Services
Share with your colleagues
Related articles.

How to write an Introduction to an academic article
Charlesworth Author Services 17/08/2020 00:00:00
Writing an Abstract: Purpose and Tips
Related webinars.

Bitesize Webinar: How to write and structure your academic article for publication- Module 3: Understand the structure of an academic paper
Charlesworth Author Services 04/03/2021 00:00:00

Bitesize Webinar: How to write and structure your academic article for publication: Module 6: Choose great titles and write strong abstracts
Charlesworth Author Services 05/03/2021 00:00:00

Bitesize Webinar: How to write and structure your academic article for publication: Module 11: Know when your article is ready for submission

Bitesize Webinar: How to write and structure your academic article for publication - Module 14: Increase your chances for publication
Charlesworth Author Services 20/04/2021 00:00:00

Authorship of academic papers
Charlesworth Author Services 11/07/2017 00:00:00

Ethics in academic publishing: Understanding ‘gift’ authorships
Charlesworth Author Services 21/11/2019 00:00:00

Who gets the CRediT? Authorship issues and fairness in academic writing
Charlesworth Author Services 15/07/2019 00:00:00
- Buy Custom Assignment
- Custom College Papers
- Buy Dissertation
- Buy Research Papers
- Buy Custom Term Papers
- Cheap Custom Term Papers
- Custom Courseworks
- Custom Thesis Papers
- Custom Expository Essays
- Custom Plagiarism Check
- Cheap Custom Essay
- Custom Argumentative Essays
- Custom Case Study
- Custom Annotated Bibliography
- Custom Book Report
- How It Works
- +1 (888) 398 0091
- Essay Samples
- Essay Topics
- Research Topics
- Uncategorized
- Writing Tips
How to Write Acknowledgement in Research Paper
December 27, 2023
In research papers, acknowledgements play a crucial role in recognizing the contributions and support of individuals or organizations who have helped in the completion of the study. They provide an opportunity to express gratitude towards people who provided guidance, resources, or encouragement during the research process.
Acknowledgements serve several important purposes. Firstly, they acknowledge the intellectual and technical support received, ensuring that credit is given where it is due. Secondly, they foster a sense of collaboration and community by recognizing the efforts of others within the research field. Furthermore, acknowledgements help to establish credibility by showcasing the individuals, institutions, or funding bodies that have contributed to the research.
The importance of acknowledgements goes beyond mere courtesy; they are a record of the network of support that exists within the research community. By recognizing the contributions of others, researchers can foster goodwill and build lasting relationships in their field. As such, it is crucial to understand the significance of acknowledgements and to include them thoughtfully and sincerely in research papers.
What to Include in the Acknowledgements
When writing acknowledgements in a research paper, it is important to consider the key elements that should be included. Here are some points to consider when deciding what to include in your acknowledgements:
- Mentors and Advisors: Acknowledge the individuals who provided guidance, advice, or supervision throughout the research process. This could include your research advisor, committee members, or other mentors who played a significant role in shaping your work.
- Funding Sources: Acknowledge any funding bodies or organizations that provided financial support for your research. This could include grants, scholarships, or fellowships.
- Research Participants: If your study involved human participants or subjects, it is important to thank them for their participation and contribution to the research. Ensure that their privacy and confidentiality are maintained by using initials or codes instead of full names.
- Collaborators and Colleagues: If you worked closely with other researchers or colleagues on the project, acknowledge their contributions and highlight their specific roles or expertise.
- Technical and Administrative Support: Recognize individuals or institutions that provided technical assistance, equipment, or administrative support during the research process.
- Family and Friends: It is common to express gratitude to loved ones for their support and understanding throughout the research journey. Although personal acknowledgements should be kept brief, acknowledging their support can be a significant gesture.
Remember to be sincere and genuine in your acknowledgements, recognizing the specific contributions and support each individual or entity provided.
Structure and Formatting of the Acknowledgements
The structure and formatting of acknowledgements in a research paper play a vital role in conveying gratitude in a clear and organized manner. Here are some guidelines to follow when structuring and formatting your acknowledgements:
- Placement: The acknowledgements section is typically placed towards the end of the paper, after the conclusion but before the references.
- Title: Use a clear and concise title such as “Acknowledgements” or “Acknowledgments” at the top of the section to indicate its purpose.
- Formatting: In general, acknowledgements are written in paragraph form. However, if you have a long list of individuals or organizations to acknowledge, you can also use a bullet-point or numbered list.
- Tone: The tone of acknowledgements should be professional and appreciative. Use a sincere and heartfelt tone to express your gratitude.
- Length: Acknowledgements should be kept concise and to the point. A few paragraphs or a short list of names and institutions are typically sufficient.
- Order: It is customary to acknowledge individuals and organizations in order of their significance and contribution to the research. Start with your most important mentors, advisors, or sponsors, and then proceed to acknowledge collaborators, research participants, and others who provided support.
- Style: Follow the formatting guidelines specified by the journal or academic institution. This may include font type, size, and spacing.
Remember, while acknowledgements provide an opportunity to express gratitude, it is important to maintain professionalism and adhere to the guidelines set by the journal or your academic institution.
Examples of Well-Written Acknowledgements
Example 1: “I would like to express my deepest gratitude to Professor [Name] for their unwavering guidance, invaluable insights, and constant support throughout this research journey. Their expertise and dedication have been instrumental in shaping this study. I would also like to thank my research committee members, [Names], for their thoughtful feedback and constructive criticism. I am grateful to [Funding Body/Institution] for providing financial support that made this research possible. Special thanks to all the participants who generously volunteered their time and contributed to this study. I would also like to acknowledge [Support Staff/Technicians] for their technical assistance. Lastly, I am indebted to my family and friends for their unwavering support, love, and encouragement throughout this endeavor.”
Example 2: “We extend our sincere appreciation to Professor [Name] for their mentorship and guidance, which have been invaluable in the completion of this research. We would like to thank [Funding Body/Institution] for their financial support, without which this study would not have been possible. We are grateful to the participants who willingly shared their experiences and expertise, providing the foundation for our research. Our gratitude also extends to our collaborators and fellow researchers who provided valuable insights and feedback throughout the project. We would like to acknowledge the technical support provided by [Support Staff/Technicians]. Lastly, we express our heartfelt thanks to our families and friends for their unwavering support and understanding.”
Remember, these examples are just a guide. It is important to personalize your acknowledgements based on the individuals and organizations that have contributed to your specific research project.
Tips for Writing Acknowledgements
- Be specific: Clearly mention the names of individuals, institutions, or organizations that have made significant contributions to your research.
- Focus on contributions: Acknowledge the specific ways in which each person or entity has contributed to your research, whether it’s through mentorship, funding, technical support, or intellectual guidance.
- Maintain professionalism: While expressing gratitude, maintain a professional tone and avoid overly personal or informal language.
- Be concise: Keep your acknowledgements brief and to the point, focusing on the most essential contributors.
- Follow guidelines: Adhere to any specific formatting guidelines provided by the journal or academic institution.
- Review and proofread: Ensure that your acknowledgements are free of spelling or grammatical errors by reviewing and proofreading them carefully.
- Express sincerity: Convey genuine appreciation and gratitude in your acknowledgements to show the true extent of your appreciation.
Remember, acknowledgements are an opportunity to show gratitude to those who have supported your research, so be sure to express your thanks sincerely and respectfully.
Common Mistakes to Avoid in Acknowledgements
- Excessive length: Avoid making your acknowledgements overly lengthy. Keep them concise and focus only on those who have made substantial contributions to your research.
- Forgetting important contributors: Take the time to think through all the individuals and organizations that have played a role in your research and ensure that you acknowledge them appropriately. Leaving out key contributors can be unintentionally dismissive.
- Being vague: Be specific when acknowledging the contributions of individuals or institutions. Avoid using generic statements that don’t adequately highlight the specific ways in which they have supported your research.
- Inclusion of personal anecdotes: While it’s important to express gratitude, acknowledgements should remain professional. Avoid including personal anecdotes or unrelated stories that distract from the purpose of the section.
- Neglecting proofreading: Treat acknowledgements with the same level of attention and care as the rest of your research paper. Proofread for spelling and grammatical errors before submitting your final document.
- Inconsistent formatting: Ensure that your acknowledgements follow the same formatting as the rest of your research paper, adhering to any journal or institution guidelines.
By avoiding these common mistakes, you can ensure that your acknowledgements accurately reflect the contributions of those involved in your research while maintaining a professional and concise tone.
Struggling with your research paper’s acknowledgement section? Our auto essay writer can make it easy, guiding you to express your gratitude flawlessly.
Sociology Research Topics Ideas
Importance of Computer in Nursing Practice Essay
History Research Paper Topics For Students
By clicking “Continue”, you agree to our terms of service and privacy policy. We’ll occasionally send you promo and account related emails.
Latest Articles
Navigating the complexities of a Document-Based Question (DBQ) essay can be daunting, especially given its unique blend of historical analysis...
An introduction speech stands as your first opportunity to connect with an audience, setting the tone for the message you...
Embarking on the journey to write a rough draft for an essay is not just a task but a pivotal...
I want to feel as happy, as your customers do, so I'd better order now
We use cookies on our website to give you the most relevant experience by remembering your preferences and repeat visits. By clicking “Accept All”, you consent to the use of ALL the cookies. However, you may visit "Cookie Settings" to provide a controlled consent.
Acknowledgments in Scientific Papers
- Published: 07 July 2023
- Volume 39 , pages 280–299, ( 2023 )
Cite this article
- Jaime A. Teixeira da Silva 1 ,
- Panagiotis Tsigaris 2 &
- Quan-Hoang Vuong 3
549 Accesses
1 Altmetric
Explore all metrics
Acknowledgements are usually a minor part of scientific papers, but they serve a very important function. This section of the manuscript is normally reserved to thank those who offered assistance, but not enough to merit authorship, funders, or any other people or organizations or artificial intelligent tools that may have in any way been directly associated either with the research reported in that study, or with the published paper. Despite this, it is not uncommon to see wide disparities in ethics and author guidelines pertaining to acknowledgements, as was observed in 45 publishing-related entities (journals, publishers, preprints, ethics organizations, open access aggregators) that were assessed in this study. Greater standardization is required, especially among members of ethics policy groups such as the Committee on Publication Ethics or the International Committee of Medical Journal Editors. Moreover, even though verification is an essential step of this process, it is difficult to achieve.
This is a preview of subscription content, log in via an institution to check access.
Access this article
Price includes VAT (Russian Federation)
Instant access to the full article PDF.
Rent this article via DeepDyve
Institutional subscriptions
Similar content being viewed by others
Do reviewers get their deserved acknowledgments from the authors of manuscripts?
Pengfei Jia, Weixi Xie, … Xianwen Wang

Scientific Electronic Library Online/SciELO: Good Practices Guide for the Enhancement of Ethics in Scientific Publication (Version 09/2018)

Publication ethics: Role and responsibility of authors
Shubha Singhal & Bhupinder Singh Kalra
“Big Science” refer to mega-projects involving very large collaborative interdisciplinary teams with moments in history of great exploration such as the Human Genome Project, the Large Hardon Collider and NASA’s international space station, to name a few. A physics paper about the Large Hardon Collider had 5154 authors [ 4 ], a classic case of hyper-authorship [ 5 ]. If those being acknowledged are seen as collaborators in research output, which they should be, then the lone wolf scholar is indeed an endangered species [ 6 , 7 ].
Cronin on page 19 states: “Who is to say whether the least significant co-author’s contributions were greater or less than those of the most helpful acknowledge?”.
In science research, there is strong evidence of division of work. Active participation to earn authorship is associated with one or more of the following tasks: analyzing the data, conceiving and designing the experiments, contributing to reagents, material and/or analysis tools, performing the experiment, and writing the paper [ 19 ]. Lack of contributorship statements reflect a lack of transparency and accountability according to Larivière et al. [ 19 ].
Some of these concepts were inferred by Cronin et al. [ 20 ], who indicated that one individual was named in 25 acknowledgements in Psychological Review , while three editors were also frequently acknowledged, the most frequent being in 20 papers. Cronin [ 3 ] highlighted multiple studies that formally analyzed the acknowledgements in journals such as Cell or Genetics , finding them to be a complex mixture of influences (personal, field-based, historical), often laced with thanks for laboratory gifts, and the need and expectation of reciprocity.
Cronin’s opinion is debatable, can an acknowledged person really be more important than a co-author?
This is based on our experience, and a thorough meta-analysis would be required to assess if being acknowledged has rewards in academic institutes across the globe.
Cronin [ 3 ] stated: “In a reward system of the academy, a mere acknowledgement, no matter how influential the acknowledged intellectual contribution is perceived to be, is not treated as equivalent to even the lowest form of citation” (p. 24).
There is a further tier system within the order of appearance of authors’ names and contribution. Evidence from PLOS journals suggested a U-shaped relationship between the order of authors’ names and contribution levels in which the first and last authors’ contributions are greater than those in the middle [ 19 ].
At the end of the abstract they concluded: “more accurate documentation of funding sources in published articles would benefit researchers, funders and journals, and enhance the reliability and usefulness of funding acknowledgements.
https://publicationethics.org/members .
http://www.icmje.org/journals-following-the-icmje-recommendations/ .
https://www.stm-assoc.org/membership/our-members/ .
https://oaspa.org/membership/members/ .
https://doaj.org/sponsors .
Listed publishers were: American Chemical Society, American Institute of Physics, American Physical Society, Cambridge University Press, Emerald, IEEE, Institute of Physics, Karger, Nature Publishing Group, Optical Society of America, Oxford University Press, Reed-Elsevier, Royal Society of Chemistry, Sage Publications, Springer, Taylor & Francis, Thieme Publishing Group, Wiley-Blackwell, and Wolters Kluwer. In the Supplementary Table, Nature Research was considered separately to Springer Nature, even though they fall under the same publisher.
https://fairsharing.org/ .
For all these aspects, a comprehensive analysis of all journals within each publisher listed in the Supplementary Table would be needed in order to clearly quantify deviations from ICMJE recommendations.
The ICMJE lists journals that claim to follow its recommendations, but the veracity of entries on those lists have not been verified, nor can the scholarly nature of those journals be proved. Publishers are not listed. Since several of the publishers listed in the Supplementary Table have hundreds or even thousands of journals, it is humanly impossible to verify the number of journals that claim to follow ICMJE guidelines, but this analysis is necessary.
Since deviation among clauses were detected in journals, even within the same publisher, there is a high likelihood that a clause might exist for one or more journals in several of the publishers with large journal fleets.
Flexibility is not necessarily a good thing because it opens up exceptions to the rule, and allows authors to interpret clauses and rules differently. This issue becomes particularly important for authors who have their papers rejected in one journal/publisher with rule/clause A, and then resubmit to another journal of another publisher with a different rule/clause B.
“Despite five decades of analysis putting forward the potential value of acknowledgments as markers of scientific capital, the literature still lacks consensus as to the value and functions of acknowledgments within the reward system of science.” (p. 2821).
“Professional editors (e.g. medical writers) engaged to prepare scientific texts and graphics, or to put research findings into a form suitable for publication, are to be listed as authors if, by virtue of these activities, they influence the weight attached to the findings and the impact of the publication. If they are only responsible for purely linguistic and editorial improvements, they are not to be listed in the byline; it is appropriate to mention them in the Acknowledgements.” (p. 4).
Bazerman C. Modern evolution of the experimental report in physics: spectroscopic articles in Physical Review , 1893–1980. Soc Stud Sci. 1984;14(2):163–96. https://doi.org/10.1177/030631284014002001 .
Article Google Scholar
de Lima Navarro P, de Amorim Machado C. An origin of citations: Darwin’s collaborators and their contributions to the Origin of Species. J Hist Biol. 2020;53(1):45–79. https://doi.org/10.1007/s10739-020-09592-8 ( corrigendum (2022): Journal of the History of Biology, 55(1), 205–206. https://doi.org/10.1007/s10739-021-09640-x ).
Cronin B. The scholar’s courtesy: the role of acknowledgement in the primary communication process. London: Taylor Graham; 1995. http://garfield.library.upenn.edu/cronin/cronin2part1.pdf ; http://garfield.library.upenn.edu/cronin/cronin2part2.pdf . Accessed 7 June 2023.
Castelvecchio D. Physics paper sets record with more than 5,000 authors. Nature. 2015. https://doi.org/10.1038/nature.2015.17567 .
Cronin B. Hyper authorship: a postmodern perversion or evidence of a structural shift in scholarly communication practices? J Am Soc Inf Sci Technol. 2001;52(7):558–69. https://doi.org/10.1002/asi.1097 .
Paul-Hus A, Díaz-Faes AA, Sainte-Marie M, Desrochers N, Costas R, Larivière V. Beyond funding: acknowledgement patterns in biomedical, natural and social sciences. PLoS ONE. 2017;12(10):e0185578. https://doi.org/10.1371/journal.pone.0185578 .
Paul-Hus A, Mongeon P, Sainte-Marie M, Larivière V. The sum of it all: revealing collaboration patterns by combining authorship and acknowledgments. J Informetr. 2017;11(1):80–7. https://doi.org/10.1016/j.joi.2016.11.005 .
Henriksen D. The rise in co-authorship in the social sciences (1980–2013). Scientometrics. 2016;107(2):455–76. https://doi.org/10.1007/s11192-016-1849-x .
Larivière V, Gingras Y, Archambault É. Canadian collaboration networks: a comparative analysis of the natural sciences, social sciences and the humanities. Scientometrics. 2006;68(3):519–33. https://doi.org/10.1007/s11192-006-0127-8 .
Wuchty S, Jones BF, Uzzi B. The increasing dominance of teams in production of knowledge. Science. 2007;316(5827):1036–9. https://doi.org/10.1126/science.1136099 .
Costas R, van Leeuwen TN. Approaching the “reward triangle”: general analysis of the presence of funding acknowledgments and “peer interactive communication” in scientific publications. J Am Soc Inf Sci Technol. 2012;63(8):1647–61. https://doi.org/10.1002/asi.22692 .
Cronin B, Weaver S. The praxis of acknowledgment: from bibliometrics to influmetrics. Rev Esp Doc Cient. 1995;18(2):172–7. https://doi.org/10.3989/redc.1995.v18.i2.654 .
International Committee of Medical Journal Editors (ICMJE). Recommendations. 2023. https://www.icmje.org/icmje-recommendations.pdf . Accessed 7 June 2023.
Teixeira da Silva JA, Tsigaris P. Human- and AI-based authorship: principles and ethics. Learn Publ. 2023. https://doi.org/10.1002/leap.1547 .
Teixeira da Silva JA. How are authors’ contributions verified in the ICMJE model? Plant Cell Rep. 2023. https://doi.org/10.1007/s00299-023-03022-9 .
Teixeira da Silva JA, Dobránszki J. Multiple authorship in scientific manuscripts: ethical challenges, ghost and guest/gift authorship, and the cultural/disciplinary perspective. Sci Eng Ethics. 2016;22(5):1457–72. https://doi.org/10.1007/s11948-015-9716-3 .
Wislar JS, Flanagin A, Fontanarosa PB, DeAngelis CD. Honorary and ghost authorship in high impact biomedical journals: a cross sectional survey. BMJ. 2011;343:d6128. https://doi.org/10.1136/bmj.d6128 .
Cislak A, Formanowicz M, Saguy T. Bias against research on gender bias. Scientometrics. 2018;115(1):189–200. https://doi.org/10.1007/s11192-018-2667-0 .
Larivière V, Desrochers N, Macaluso B, Mongeon P, Paul-Hus A, Sugimoto CR. Contributorship and division of labor in knowledge production. Soc Stud Sci. 2016;46(3):417–35. https://doi.org/10.1177/0306312716650046 .
Cronin B, Shaw D, La Barre K. A cast of thousands: coauthorship and subauthorship collaboration in the 20th century as manifested in the scholarly journal literature of psychology and philosophy. J Am Soc Inf Sci Technol. 2003;54(9):855–71. https://doi.org/10.1002/asi.10278 .
Song M, Kang K-Y, Timakum T, Zhang X. Examining influential factors for acknowledgements classification using supervised learning. PLoS ONE. 2020;15(2):e0228928. https://doi.org/10.1371/journal.pone.0228928 .
Tsigaris P, Teixeira da Silva JA. The role of ChatGPT in scholarly editing and publishing. Eur Sci Ed. 2023;49:e101121. https://doi.org/10.3897/ese.2023.e101121 .
Cronin B, Franks S. Trading cultures: resources mobilization and service rendering in the life sciences as revealed in the journal articles’ paratext. J Am Soc Inf Sci Technol. 2006;57(14):1909–18. https://doi.org/10.1002/asi.20407 .
Giles CL, Councill IG. Who gets acknowledged: measuring scientific contributions through automatic acknowledgment indexing. Proc Natl Acad Sci USA. 2004;101(51):17599–604. https://doi.org/10.1073/pnas.0407743101 .
Baas J, Schotten M, Plume A, Côté G, Karimi R. Scopus as a curated, high-quality bibliometric data source for academic research in quantitative science studies. Quant Sci Stud. 2020;1(1):377–86. https://doi.org/10.1162/qss_a_00019 .
Brown R. How scholars credit editors in their acknowledgements. J Sch Publ. 2009;40(4):384–98. https://doi.org/10.3138/jsp.40.4.384 .
Tiew W-S, Sen BK. Acknowledgement patterns in research articles: a bibliometric study based on Journal of Natural Rubber Research 1986–1997. Malays J Libr Inf Sci. 2002;7(1):43–56.
Google Scholar
Salager-Meyer F, AlcarazAriza MA, PabónBerbesí M. “Backstage” solidarity in Spanish- and English-written medical research papers: publication context and the acknowledgment paratext. J Am Soc Inf Sci Technol. 2009;60(2):307–17. https://doi.org/10.1002/asi.20981 ( correction (2014): https://doi.org/10.1002/asi.23402 ).
Liu W, Tang L, Hu G. Funding information in Web of Science: an updated overview. Scientometrics. 2020;122(3):1509–24. https://doi.org/10.1007/s11192-020-03362-3 .
Tang L, Hu G, Liu W. Funding acknowledgment analysis: queries and caveats. J Am Soc Inf Sci. 2017;68(3):790–4. https://doi.org/10.1002/asi.23713 .
Paul-Hus A, Desrochers N, Costas R. Characterization, description, and considerations for the use of funding acknowledgment data in Web of Science. Scientometrics. 2016;108(1):167–82. https://doi.org/10.1007/s11192-016-1953-y .
Álvarez-Bornstein B, Morillo F, Bordons M. Funding acknowledgments in the Web of Science: completeness and accuracy of collected data. Scientometrics. 2017;112(3):1793–812. https://doi.org/10.1007/s11192-017-2453-4 .
Méndez DI, Alcaraz MA. Exploring acknowledgement practices in English-medium astrophysics research papers: Implications on authorship. Revista de Lenguas para Fines Específicos. 2015;21(1):132–59. https://doi.org/10.20420/rlfe.2015.0001 .
Díaz-Faes AA, Bordóns M. Acknowledgments in scientific publications: presence in Spanish science and text patterns across disciplines. J Am Soc Inf Sci. 2014;65(9):1834–49. https://doi.org/10.1002/asi.23081 .
Kassirer JP, Angell M. On authorship and acknowledgments. N Engl J Med. 1991;325:1510–2. https://doi.org/10.1056/NEJM199111213252112 .
Teixeira da Silva JA, Dobránszki J. How authorship is defined by multiple publishing organizations and STM publishers. Account Res. 2016;16(2):97–122. https://doi.org/10.1080/08989621.2015.1047927 .
Teixeira da Silva JA, Dobránszki J. Notices and policies for retractions, expressions of concern, errata and corrigenda: their importance, content, and context. Sci Eng Ethics. 2017;23(2):521–54. https://doi.org/10.1007/s11948-016-9769-y .
Teixeira da Silva JA, Dobránszki J. Preprint policies among 14 academic publishers. J Acad Librariansh. 2019;45(2):162–70. https://doi.org/10.1016/j.acalib.2019.02.009 .
Larivière V, Haustein S, Mongeon P. The oligopoly of academic publishers in the digital era. PLoS ONE. 2015;10(6):e0127502. https://doi.org/10.1371/journal.pone.0127502 .
Allen C, Mehler DMA. Open science challenges, benefits and tips in early career and beyond. PLoS Biol. 2019;17(5):e3000246. https://doi.org/10.1371/journal.pbio.3000246 .
Flanagin A, Fontanarosa PB, Bauchner H. Preprints involving medical research—do the benefits outweigh the challenges? JAMA. 2020;324(18):1840–3. https://doi.org/10.1001/jama.2020.20674 .
PhilPapers. About PhilPapers. PhilPapers; 2021. https://philpapers.org/help/about.html . Accessed 7 June 2023.
Sansone S-A, McQuilton P, Rocca-Serra P, Gonzalez-Beltran A, Izzo M, Lister AL, Thurston M. FAIRsharing as a community approach to standards, repositories and policies. Nat Biotechnol. 2019;37:35. https://doi.org/10.1038/s41587-019-0080-8 .
Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, Bouwman J, Mons B. The FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3(1):1–9. https://doi.org/10.1038/sdata.2016.18 .
Wilkinson MD, Sansone SA, Schultes E, Doorn P, da Silva Santos LOB, Dumontier M. A design framework and exemplar metrics for FAIRness. Sci Data. 2018;5(1):1–4. https://doi.org/10.1038/sdata.2018.118 .
Teixeira da Silva JA. The ICMJE recommendations: challenges in fortifying publishing integrity. Ir J Med Sci. 2020;189(4):1179–81. https://doi.org/10.1007/s11845-020-02227-1 .
Fontanarosa P, Bauchner H, Flanagin A. Authorship and team science. J Am Med Assoc. 2017;318(24):2433–7. https://doi.org/10.1001/jama.2017.19341 .
Teixeira da Silva JA. Outsourced English revision, editing, publication consultation and integrity services should be acknowledged in an academic paper. J Nanopart Res. 2021;23(4):81. https://doi.org/10.1007/s11051-021-05199-0 .
Teixeira da Silva JA. Should (religious) deities be acknowledged? Asian Aust J Plant Sci Biotechnol. 2013;7(1):122–3.
Jamieson KH. Crisis or self-correction: rethinking media narratives about the well-being of science. Proc Natl Acad Sci USA. 2018;115(11):2620–7. https://doi.org/10.1073/pnas.1708276114 .
Resnik DB, Tyler AM, Black JR, Kissling G. Authorship policies of scientific journals. J Med Ethics. 2016;42(3):199–202. https://doi.org/10.1136/medethics-2015-103171 .
Teixeira da Silva JA. Challenges to open peer review. Online Inf Rev. 2019;43(2):197–200. https://doi.org/10.1108/OIR-04-2018-0139 .
Teixeira da Silva JA, Dobránszki J. Editors moving forward: stick to academic basics, maximize transparency and respect, and enforce the rules. Recent Prog Med. 2018;109(5):263–6. https://doi.org/10.1701/2902.29244 .
Desrochers N, Paul-Hus A, Pecoskie J. Five decades of gratitude: a meta-synthesis of acknowledgments research. J Am Soc Inf Sci. 2017;68(12):2821–33. https://doi.org/10.1002/asi.23903 .
Burrough-Boenisch J. Do freelance editors for academic and scientific researchers seek acknowledgement? Findings from a cross-sectional study. Eur Sci Ed. 2019;45(2):32–8. https://doi.org/10.20316/ESE.2019.45.18019 .
Matarese V, Shashok K. Transparent attribution of contributions to research: aligning guidelines to real-life practices. Publications. 2019;7(2):24. https://doi.org/10.3390/publications7020024 .
Matarese V, Shashok K. Acknowledging editing and translation: a pending issue in accountability. Account Res. 2020;27(4):238–9. https://doi.org/10.1080/08989621.2020.1737525 .
Hess CW, Brückner C, Kaiser T, Mauron A, Wahli W, Wenzel UJ, Salathé M. Authorship in scientific publications: analysis and recommendations. Swiss Med Wkly. 2015;145:w14108. https://doi.org/10.4414/smw.2015.14108 .
Mlinarić A, Horvat M, Smolčić VŠ. Dealing with the positive publication bias: why you should really publish your negative results. Biochem Med. 2017;27(3):030201. https://doi.org/10.11613/BM.2017.030201 .
Download references
Acknowledgments
The authors acknowledge the feedback of an anonymous reviewer as well as the journal’s Editor-in-Chief.
Author information
Authors and affiliations.
Independent Researcher, Ikenobe 3011-2, Miki-cho, Kagawa-ken, 761-0799, Japan
Jaime A. Teixeira da Silva
Department of Economics, Thompson Rivers University, Kamloops, BC, V2C 0C8, Canada
Panagiotis Tsigaris
Centre for Interdisciplinary Social Research, Phenikaa University, Ha Dong District, Hanoi, 100803, Viet Nam
Quan-Hoang Vuong
You can also search for this author in PubMed Google Scholar
Contributions
The authors, who are co-corresponding authors, contributed equally to the intellectual discussion underlying this paper, literature exploration, writing, reviews and editing.
Corresponding author
Correspondence to Jaime A. Teixeira da Silva .
Ethics declarations
- Conflict of interest
The authors declare no conflicts of interest of relevance to this topic.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (XLSX 17 KB)
Rights and permissions.
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Reprints and permissions
About this article
Teixeira da Silva, J.A., Tsigaris, P. & Vuong, QH. Acknowledgments in Scientific Papers. Pub Res Q 39 , 280–299 (2023). https://doi.org/10.1007/s12109-023-09955-z
Download citation
Accepted : 27 June 2023
Published : 07 July 2023
Issue Date : September 2023
DOI : https://doi.org/10.1007/s12109-023-09955-z
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Accountability
- Editorial responsibility
- Peer review
- Post-publication peer review
- Quality control
- Transparency
- Find a journal
- Publish with us
- Track your research
An official website of the United States government
The .gov means it’s official. Federal government websites often end in .gov or .mil. Before sharing sensitive information, make sure you’re on a federal government site.
The site is secure. The https:// ensures that you are connecting to the official website and that any information you provide is encrypted and transmitted securely.
- Publications
- Account settings
Preview improvements coming to the PMC website in October 2024. Learn More or Try it out now .
- Advanced Search
- Journal List

Acknowledgements are not just thank you notes: A qualitative analysis of acknowledgements content in scientific articles and reviews published in 2015
Adèle paul-hus.
École de bibliothéconomie et des sciences de l'information, Université de Montréal, Downtown Station, Montreal, Quebec, Canada
Nadine Desrochers
Associated data.
Restrictions apply to the availability of the acknowledgement data, which is used under license from Clarivate Analytics. Readers can contact Clarivate Analytics at the following URL: http://clarivate.com/scientific-and-academic-research/research-discovery/web-of-science/ . References for the acknowledgement excerpts used are available in S1 Table .
Acknowledgements in scientific articles can be described as miscellaneous, their content ranging from pre-formulated financial disclosure statements to personal testimonies of gratitude. To improve understanding of the context and various uses of expressions found in acknowledgements, this study analyses their content qualitatively. The most frequent noun phrases from a Web of Science acknowledgements corpus were analysed to generate 13 categories. When 3,754 acknowledgement sentences were manually coded into the categories, three distinct axes emerged: the contributions, the disclaimers, and the authorial voice. Acknowledgements constitute a space where authors can detail the division of labour within collaborators of a research project. Results also show the importance of disclaimers as part of the current scholarly communication apparatus, an aspect which was not highlighted by previous analyses and typologies of acknowledgements. Alongside formal disclaimers and acknowledgements of various contributions, there seems to remain a need for a more personal space where the authors can speak for themselves, in their own name, on matters they judge worth mentioning.
Introduction
The idea of using acknowledgements as a source for bibliometric indicators has been surrounding their study since the 1990s. In 1991, Cronin was already asking, “why are acknowledgement counts excluded from formal assessments of individual merit or influence, such as tenure review?” ([ 1 ]: p. 236). In 1995, Cronin and Weaver were encouraging the development of an Acknowledgement Index, based on the model of the Science Citation Index [ 2 ]. Almost two decades later, Costas and van Leeuwen [ 3 ] suggested that it was perhaps time “to employ this sort of tool to facilitate development of the so-called ‘influmetrics’” ([ 3 ]: p. 1659). For their part, Díaz-Faes and Bordons [ 4 ] highlighted that the inclusion of acknowledgement information in the Web of Science (WoS) was offering new avenues to study collaboration in science, going beyond traditional bibliometric indicators. McCain [ 5 ] went further and assessed the feasibility of a formal Personal Acknowledgements Index. And yet, despite decades of studies positioning acknowledgements alongside citations and authorship in what Cronin called the “reward triangle” [ 6 ], the consideration of acknowledgements as an indicator of scientific credit has not materialized and, at best, remains a proposal at the exploratory stage, or even simply a rhetorical idea (see [ 7 ] for a meta-synthesis of this literature).
At the same time, many studies have used funding-related indicators based on acknowledgement data (e.g. [ 8 – 11 ]). In fact, acknowledgement studies can no longer be separated from the financial aspect of scientific research. In 2008, WoS started to collect and index funding sources found in the acknowledgements of scientific papers. These new data were added by WoS in response to many funding bodies’ requirement to acknowledge the sources supporting research. Since then, large-scale acknowledgement data have been used as a bibliometric tool to follow the money trail of research and funding-related analyses have become a dominant trend in recent acknowledgement literature [ 7 ]. To this day, acknowledgements have been more closely related to funding indicators than to any other kind of scientific credit indicators.
The literature also underlines the elusive nature of acknowledgements, pointing to their form and tone, which have been described as sometimes flowery, personal, and even manipulative:
- Acknowledgements are permeated by hyperbole, effusiveness, overstatement, and exaggeration. ([ 12 ]: p. 64)
- Acknowledgements have been discussed as a form of patronage in scholarly communication, where the reality of the past may be purposefully glossed over and where the author could be looking toward the possibility of receiving future favours. ([ 13 ]: p. 4)
Furthermore, several studies mention the lack of standardization of acknowledgements as one important limitation hindering their analyses:
- The format of acknowledgement varies from field to field and from journal to journal. As noted, persons and institutional sources may be listed in the methods and materials section of an article or explicitly thanked in an acknowledgement section. ([ 14 ]: p. 506)
- Since there are no established formats for acknowledgements in papers, as there are for citations, expressions of gratitude vary greatly and sometimes it was difficult to identify the correct type of support, and even more difficult, the correct funding organization. ([ 15 ]: p. 238)
- The first source of simple error may arise through the misspelling of the names of funding bodies and potentially the names of grants and grant codes […]. A second difficulty will be that researchers will not correctly remember the funding bodies and grants that they used to support the research. ([ 16 ]: p. 368–369)
Acknowledgements may thus contain formally required statements of gratitude but have also been used as personal spaces of authorial expression, and as such, acknowledgement texts have been analysed as a genre per se. Several discourse and linguistic analyses have studied acknowledgements found in dissertations, theses, monographies, and research articles (e.g. [ 17 – 19 ]).
Acknowledgements analyses have also led to numerous typologies or classifications of the contributions acknowledged in scientific publications. In 1972, Mackintosh [ 20 ] proposed the first qualitative content analysis of acknowledgements based on a typology of the three main types of “services” acknowledged in scientific papers: facilities , access to data , and help of individuals . Twenty years later, McCain [ 14 ] offered a finer typology of acknowledgements, using five categories: access to research-related information , access to unpublished results and data , peer interactive communication , technical assistance , and manuscript preparation . The same year, Cronin introduced his first version of a six-part typology of acknowledgements ( paymaster , moral support , dogsbody , technical , prime mover , and trusted assessor ) which was created before encountering Mackintosh’s 1972 and McCain’s 1991 work [ 1 , 21 ]. Subsequent versions of this typology—developed with different collaborators through the years (namely McKenzie, Rubio and Weaver(-Wozniak))—include the peer interactive communication category borrowed from McCain [ 14 ] alongside moral support , access (to resources, materials and infrastructure), clerical support , technical support , and financial support [ 2 , 22 – 24 ]. Cronin’s model has since been adopted, adapted, and augmented in several studies (e.g. [ 25 – 30 ].
More recently, Giles and Councill [ 31 ] used natural language processing to extract named entities from more than 180,000 acknowledgements published in computer science research papers. In their content analysis, the most frequently acknowledged entities are classified into four categories: funding agencies , corporations , universities and individuals . Other studies have analysed the content of acknowledgements focusing on funding bodies and classifying them by sectors and subsectors (e.g. [ 10 , 32 – 35 ]).
Finally, linguistic studies have also used classifications of acknowledgements, focusing on the structure and patterns of dissertation acknowledgement texts (e.g. [ 18 , 36 – 40 ]) and on the socio-pragmatic construction of acknowledgements found in research articles and academic books [ 19 , 41 – 43 ].
Typologies and classifications aim to describe and categorize the content of acknowledgements in a synthetic manner. However, these taxonomies are based on small-scale samples of acknowledgements, the only exception being the work of Giles and Councill [ 31 ] which focused solely on named entities. More recently, a large-scale multidisciplinary analysis of acknowledgement texts was published by the authors and collaborators in PLOS One [ 44 ]. This analysis of acknowledgements from more than one million articles and reviews published in 2015, highlighted important variations in the practices of acknowledging. Focusing on the 214 most frequent noun phrases of that corpus, the study showed that acknowledgement practices truly do vary across disciplines. Noun phrases referring to technical support appeared more frequently in natural sciences while noun phrases related to peers (colleagues, editors and reviewers) were more frequent in earth and space, professional fields, and social sciences. Noun phrases referring to logistics and fieldwork-related tasks appeared prominently in biology. Pre-formulated statements used in the context of conflict of interest or responsibility disclosures were more frequently found in acknowledgements from clinical medicine, health, and psychology. However, this analysis also led to further questions concerning the interpretation of these noun phrases in their original context. Findings from this study showed that acknowledgements are not limited to credit attribution and that the numerous taxonomies and classifications found in the literature do not account for the current acknowledgement practices where pre-formulated statements of financial assistance and conflict of interest disclosures appear to be frequent [ 44 ]. Conclusions from this study raise further questions because these pre-formulated statements could have an influence on large-scale analyses that use automated linguistic methods, thus calling for a qualitative analysis of acknowledgements in the context of their use.
Objective and research questions
To improve understanding of the context and various uses of expressions found in acknowledgements, this study proposes to analyse their content qualitatively. More specifically, this study aims at answering the following research questions:
- In which contexts are specific expressions used?
- Do the contexts and meanings vary by discipline?
- What does a qualitative analysis reveal in terms of offering avenues for a more contextualized use of acknowledgements in large-scale studies?
Data and methods
Data for this study were retrieved from WoS’s Science Citation Index Expanded (SCI-E) and Social Sciences Citation Index (SSCI), which both include funding acknowledgement data. It bears repeating that acknowledgments are collected and indexed by WoS only if they include funding source information [ 45 ]. Access to WoS data in a relational database format was provided by the Observatoire des sciences et des technologies ( http://www.ost.uqam.ca ). The full text of acknowledgements from all 2015 articles and reviews indexed in the SCI-E and the SSCI were extracted. The original corpus includes a total of 1,009,411 acknowledgements for as many papers.
In a previous analysis, we identified the 214 most frequent noun phrases of that corpus of acknowledgement using natural language processing [ 44 ]. For the purpose the present qualitative analysis, these 214 noun phrases were reduced to single words (e.g. “technical assistance” was reduced to “technical” and “assistance”) and redundant words were excluded, for a final corpus of 154 single words. Each single word could therefore be found in context, no matter its proximity to other single words; this offered us the possibility to code various types of occurrences of each word, whether it was part of a noun phrase or not.
The coding was done in two steps. First, an initial codebook was established inductively by one researcher to classify each of the 154 words and revised by a second researcher. All words were then coded by both researchers and their work was reconciled through “negotiated agreement” ([ 46 ]: p. 305, see also [ 47 , 48 ]). Second, 20 words were selected from the corpus of 154 words by purposeful sampling, where cases for study are selected because “they offer useful manifestations of the phenomenon of interest” ([ 49 ]: p. 40). Selection of the words included in the final sample was based on the quantitative analysis findings [ 44 ], which highlighted the potential importance of pre-formulated statements such as “The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript” (ut 000367510900041). Special attention was given to the words frequently used in those statements (e.g. analysis, collection, design, preparation). Sampling decisions were also oriented towards potential polysemous words which could lead to different contextual meanings (e.g. “assistance”). The 20 words of the final sample were coded within the context of their original sentences, extracted from acknowledgements. Words were thus used as a seed to refer back to full acknowledgement sentences.
The coding process entails data reduction where the many meanings of a sentence must be reduced or summarized under one main category [ 50 ] in order to reflect a practice or a phenomenon on a humanly manageable scale. The principles of saturation and qualitative sampling, whereby the sample is “conceptually representative of the set of all possible units” ([ 51 ]: p. 84), ensures that the phenomenon is reflected in its full complexity. Therefore, acknowledgements were stratified by discipline to reflect potentially different disciplinary uses of a word. Coding was then performed on this sample of 20 words within their original acknowledgement contexts, using the sentence as the unit of analysis and adapting the codebook in an iterative manner as finer meanings emerged.
The final codebook is composed of 13 categories, presented in Table 1 . The coding was done by one researcher and guided by the question, “in which context is this word used?” One category was selected for each sentence coded, aiming at qualifying the context in which a word is used. Each word of the sample was coded in a minimum of 15 original sentences per discipline, for all 12 disciplines, resulting in a total of 3,754 sentences coded. Results are reported in “thick description” using sufficient descriptions and quotations to allow “thick interpretation”, which means connecting individual cases to the larger context without going into trivial details ([ 49 ]: p. 503).
The results of the coding process are summarized in Table 2 which presents, for each word of the sample, the percentage of all the occurrences attributed to a specific category. The analysis reveals the importance of three distinct axes: the contributions, the disclaimers, and the authorial voice. Moreover, disciplinary patterns bring another layer of analysis as divergent uses of the coded words emerge.
Words are presented in the table in descending order of their frequency in the corpus.
* “Other” regroups the following categories: Supervision and Management, Combination, and Vague or other.
Contributions
Acknowledgements constitute a space where authors can detail “who has done what” during the research process. Most often, authors use this space to thank colleagues that contributed to the research, as in the following example: “The authors thank Colleen Dalton and four anonymous reviewers for their helpful comments that improved the manuscript. We thank Fan-Chi Lin for providing FTAN measurements for comparison, and Anna Foster, Jiayi Xie and Goran Ekstrom for informative discussion.” (ut 000355321800013; earth and space). However, in some cases acknowledgements can also include contributorship statements from the authors in order to reflect the distribution of labour: “A.P., V.M. and V.P were involved in writing the manuscript. A.B.G and Y.A.K. were responsible for conception of the idea” (ut 000365808000014; clinical medicine).
The categories peer communication, investigation and analysis, materials and resources, and writing refer to specific types of contribution to research. These categories, taken together, represent half (50%) of the sample coded, confirming the importance of the contributions axis within the acknowledgements’ context. Moreover, some words are used most often to refer to specific categories of contribution, such as “access” which is used mainly in the category materials and resources (70% of the occurrences coded), “discussion” which is almost exclusively associated to the peer communication category (98% of the occurrences coded), and “assistance”, “experiment”, “help”, and “measurement”, which are all mainly associated to the category investigation and analysis (more than 60% of the occurrences coded).
Disclaimers
Acknowledgements are not necessarily thank-you notes or recognition of responsibility. Financial disclosure, conflict of interest, disclaimer, and ethics account for more than 40% of the sample coded. In fact, the categories financial disclosure and disclaimer are among the most frequent in the sample, accounting respectively for 22% and 18% of all occurrences coded. The words “analysis”, “collection”, “decision”, “design”, “interpretation”, “preparation”, and “writing”, which could all seemingly refer to types of contributions, were in fact used in the context of responsibility statements in a substantial share of the cases analysed. Moreover, the words “decision”, “design” and “interpretation” also are mostly found in those kinds of responsibility disclaimers (in respectively 65%, 55% and 61% of the occurrences coded for these specific words).
Non-responsibility statements of funding bodies are the most frequent disclaimers. The following example presents a typical statement: “The funding source had no role in the design of the study, the analysis and interpretation of the data or the writing of, nor the decision to publish the manuscript.” (ut 000352854700010). However, we found declarations of non-responsibility for other types of contributors regarding some part of a research project, as in the following sentence: “The data collectors have no responsibility over the analysis and interpretations presented in this study.” (ut 000349266800011). Furthermore, disclaimers are not always non-responsibility statements and can, on the contrary, disclose the specific responsibility of an organization, such as: “This study was funded by Xi'an Janssen Pharmaceutical Ltd (Beijing, People's Republic of China) who was responsible for study design and data collection, analysis, and interpretation.” (ut 000356594900001).
Contributions and disclaimers crossovers
In many cases, the disciplinary stratification provided a further level of analysis. The words “analysis”, “assistance”, and “code” present clear disciplinary patterns where the coding highlights the distinction between the two main contextual uses: the contributions axis and the disclaimers axis. For instance, the word “analysis” is used primarily in the sample to describe an investigation and analysis type of contribution: “We are grateful to Nahoko Adachi for her help in conducting the statistical analysis” (ut 000353959400005; psychology). However, for biomedical research, clinical medicine, and health, “analysis” is used mainly within the category disclaimer (example: “The funding agencies did not have any role in study design, data collection and analysis, decision to publish, or preparation of the manuscript” [ut 000346498800018; clinical medicine]). Mathematics is a divergent discipline, where the dominant category for “analysis” is financial disclosure, as exemplified by the following sentence: “This work was supported by the International Max-Planck Research School, 'Analysis, Design and Optimization in Chemical and Bio-chemical Process Engineering', Otto-von-Guericke-Universitat Magdeburg” (ut 000362588800005; mathematics).
Similarly, the word “assistance” is generally used across disciplines to describe a contribution pertaining to the category investigation and analysis (example: “The authors thank S. Watmough and K. Finder for assistance with field sampling at Dorset, and A. McDonough for assistance with the classification of plant species” [ut 000347756900044; earth and space]), except in engineering and technology and in mathematics where “assistance” is used to disclose financial help (financial disclosure) in the majority of the cases examined, as in this sentence: “The financial assistance of the National Research Foundation (NRF grant: Unlocking the future- FA2007043000003) towards this research is hereby acknowledged” (ut 000350024900008; mathematics).
Two distinct contextual uses emerge for the word “code”: it is found most often within the disclaimers axis (financial disclosure category) in biology, biomedical research, chemistry, health, psychology and social sciences (example: “The research (project code: TSY-11-3820) was supported by the Research Fund of Erciyes University” [ut 000363704000011; biology]) while it is used to describe a specific contribution (investigation and analysis category) in the majority of the cases studied in earth and space, engineering and technology, mathematics, physics and professional fields (example: “We thank Prof. D. Karaboga and Dr. B. Basturk for providing their excellent ABC MATLAB codes to implement this research” [ut 000361400900022; earth and space]).
In the case of the word “review”, the coding process also highlights two dominant uses, varying with the discipline: in biology, biomedical research, earth and space, mathematics, physics, and in the professional fields, “review” is used primarily to describe some part of the peer communication process (peer communication category), as in the following example: “We would like to express our gratitude to the anonymous referee for his or her careful review and insightful comments, in particular, for pointing out a simple proof of Lemma 1.8.” (ut 000347714700003; engineering and technology). However, in clinical medicine, a different use is made of the word “review,” mainly to refer to the document per se (dissemination category), as in this example: “We are grateful to Dr. Mozzetta for critically reading the manuscript and all members of the lab for stimulating discussions during the preparation of this review” (ut 000352374400001; clinical medicine). For all the remaining disciplines (chemistry, health, psychology, and social sciences), both categories (peer communication and dissemination) appear frequently.
The word “data” also presents distinct disciplinary patterns in the sample coded. “Data” is used mainly within the contributions axis (materials and resources category) in biology, clinical medicine, earth and space, engineering and technology, and social sciences (example: “The authors thank Chesapeake Energy for providing access to the VSP data we used” [ut 000364362900035; earth and space]). Moreover, the word “data” refers to a task within the investigation and analysis category in an important share of the cases coded in chemistry, physics, professional fields, and psychology (example: “We thank all graduate research assistants who helped with data collection” [ut 000348882900009; psychology]). However, “data” is mainly found within the disclaimers axis in clinical medicine and health (disclaimer category) as in the following example: “The funding agencies had no role in the study design, data collection and analysis, the decision to publish or preparation of the manuscript” [ut 000345586900003; clinical medicine].
Authorial voice
Although details of contributions and various disclaimers represent a substantive share of their content, acknowledgements also constitute a space for personal testimony. Notwithstanding the expectations of funders and ethical considerations, acknowledgements remain the subjective presentation of researchers’ practices and of research contexts. The authors are the voice of the acknowledgements and as such, the word “author” is one of the most frequent with more than 339,000 occurrences in our dataset. Moreover, even when the word “author” is absent, the concept is not. In fact, the authorial voice cannot be reduced to a single category, because it pervades the acknowledgements whether the authors speak in the first or third persons:
- “ I would like to thank Iliana Flores, Amy Harrison, and Shannon Kahlden for their help with data collection.” (ut 000361977300090)
- “ We would also like to thank two anonymous reviewers for the contributions to this manuscript.” (ut 000364777400031)
- “Also, our thanks go to Mr Vit Hanousek who designed an original computer tool suitable for making all the above-discussed measurements.” (ut 000346267600010)
- “The authors declare that they have no competing interests.” (ut 000369908800022)
- “The authors wish to express their appreciation to the National Iranian Copper Industry Company (NICICO) for funding this work.” (ut 000344595900005)
- “Schuster is profoundly grateful to all the families who hosted her but especially Hasidullah, his wife, son and grandson who were unfailingly patient and kind with the strange cuckoo in their nest and to the Leverhulme Trust for funding her time in Afghanistan.” (ut 000350285300006)
- “This review is dedicated to the memory of my father who was a source of inspiration.” (ut 000349637500005)
Furthermore, as exemplified by the cases presented above, the varied nature of the testimonies found in acknowledgements underlines a need for a “free space” within research publications. Alongside formal disclaimers and acknowledgements of various contributions, authors seem to require a more personal space where they can speak for themselves, in their own name, on matters they judge worth mentioning.
Discussion and conclusion
In the last decades, acknowledgements have become a “constitutive element of academic writing” ([ 52 ]: p. 160). However, the acknowledgement section is not a mandatory part of a scientific article and its content could certainly be described as miscellaneous, ranging from pre-formulated financial disclosure statements to personal testimonies of gratitude. Moreover, acknowledgements’ content and practices have evolved over time, just as citations and authorship attribution practices have changed following the transformations that are affecting the whole reward system of science [ 53 ].
Typologies and classifications of acknowledgements have been a consistent topic in the acknowledgement literature [ 7 ]. Most of these typologies and classifications revolve around the contributions axis of acknowledgements, focusing on “who gets thanked for what” and “what types of contributions are acknowledged”. This qualitative analysis of acknowledgement content confirms the importance of the contributions axis: acknowledgements are indeed still a space where authors can detail the division of labour within all collaborators of a research project. Our findings also reveal the importance of disclaimers as part of the current scholarly communication apparatus, an aspect which was not highlighted by previous analyses and typologies.
It should be noted that our analysis was restricted to a corpus of single words, sampled from noun phrases identified by correspondence analysis [ 44 ]. Further research could now seek to recombine those single words into noun phrases that present variations in meaning around a common concept, such as “assistance” (e.g. “technical assistance” and “financial assistance”). Furthermore, our coding of acknowledgement sentences was done using mutually exclusive categories, an epistemological choice. Given the fact that sentences can perform more than one kind of action, another avenue would be to use open coding and place occurrences in non-exclusive, mutually complementary categories.
Our qualitative results show that caution should be used when working with acknowledgement data. Large-scale acknowledgement data are limited to funded research, given that in the two main bibliographic databases, Web of Science and Scopus, acknowledgements are collected with the intended objective of identifying funding sponsors and tracking funded research [ 54 , 55 ]. The indexation of acknowledgements are thus limited to acknowledgements that contain some kind of funding information; this could in turn induce a potential bias toward funding-related aspects within acknowledgements’ content [ 45 ]. This indexation bias could then, at least in part, explain the importance of funding disclosures in the dataset analysed here, but also elsewhere in large-scale studies.
Yet, our findings show that acknowledgements cannot be described as having one single and homogeneous purpose; they can include expected, if not imposed, acknowledgement of financial resources as well as infrastructure alongside very personal testimonies of gratitude, all at the same time, as the following excerpt exemplifies: “Data presented herein were obtained at the W. M. Keck Observatory, which is operated as a scientific partnership among the California Institute of Technology, the University of California, and the National Aeronautics and Space Administration. […]. The authors wish to extend special thanks to those of Hawaiian ancestry, on whose sacred mountain we are privileged to be guests. Without their generous hospitality, the observations would not have been possible” (ut 000363471600015). On rare occasions, personal matters discussed in the acknowledgements become the center of attention, such as when an author proposed to his girlfriend in the acknowledgement of a paper: “C.M.B. would specifically like to highlight the ongoing and unwavering support of Lorna O’Brien. Lorna, will you marry me?” [ 56 ]. This particular paper was covered by many news outlets and online media sites when it was published, ranking in the 20 th position of the Altmetrics Top100 ranking for the year 2015. Such a case highlights the potential unexpected effect an acknowledgement can have on the visibility of a paper.
Clearly delimited and dedicated spaces for funding information, conflict of interest disclosures and contributorship statements are already implemented in some scientific journals (e.g. PLOS One , The Lancet , Science ). Nonetheless, such examples are far from the norm at the moment. In light of our findings, if an effort of standardization of acknowledgements is to be made, acknowledgements should at least include three main sections: ethics of research (financial disclosure, conflict of interest and responsibility disclaimers), contributions made to research, and personal testimony. These three indexation fields would, in turn, allow large-scale analysis of acknowledgements without the equivocality that currently characterizes these texts, yet without narrowing the space left for the authorial voice. The question remains as to whether there is a real wish within the scientific community to delineate such acknowledgement sections; if not, acknowledgement data are likely destined to remain simple tracking devices for science funding, the contributions and the authorial voices lost in large-scale analyses of scientific credit.
Supporting information
References are presented in order of in-text appearance.
Acknowledgments
The authors would like to thank Vincent Larivière for his comments and the three anonymous reviewers for their insightful suggestions and careful reading of the manuscript. This research was supported by the Social Sciences and Humanities Research Council of Canada: Joseph-Armand Bombardier CGS Doctoral Scholarships (Paul-Hus) and, Insight Development [grant number 430-2014-0617] (Desrochers).
Funding Statement
APH was supported by the Social Sciences and Humanities Research Council of Canada ( http://www.sshrc-crsh.gc.ca/ ): Joseph-Armand Bombardier CGS Doctoral Scholarships. ND was supported the Social Sciences and Humanities Research Council of Canada ( http://www.sshrc-crsh.gc.ca/ ): Insight Development [grant number 430-2014-0617]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data Availability
Publisher's Note: The article involves the independent analysis of data from publications in PLOS ONE. PLOS ONE staff had no knowledge or involvement in the study design, funding, execution or manuscript preparation. The evaluation and editorial decision for this manuscript have been managed by an Academic Editor independent of PLOS ONE staff, per our standard editorial process. The findings and conclusions reported in this article are strictly those of the author(s).
Acknowledgement Examples
Examples of Acknowledgement for Project, Thesis and Assignments
Acknowledgement For Research Paper
Acknowledgement is the process of formally recognizing someone’s effort and thanking them for it. When it comes to research papers, acknowledgements are a way of giving credit where credit is due. They can be a great way to show your appreciation for all the help and support you’ve received while writing your paper.
If you’re wondering how to write acknowledgement for research paper, you’re in luck. In this article, I’ll be providing examples of acknowledgements for research paper.
Acknowledgement Writing Tips For Research Paper
It is always nice to be able to look back on a research project and see all of the people who made it possible. Acknowledgements are one way to do that.
Here are some tips for writing acknowledgements for research papers:
First, make sure to include all of the people who directly helped with your research. This includes your supervisor, research assistants, and anyone else who played a role in your project.
Second, don’t forget to acknowledgements the financial support that made your research possible. This could include funding from your university, government grants, or private donors.
Third, you can also use your acknowledgement to thank anyone who provided moral support during your project. This could include family, friends, or fellow researchers.
Fourth, if you want to, you can also use your acknowledgement to thank the people who inspired your research. This could be other researchers, writers, or public figures.
Finally, make sure to proofread your acknowledgement carefully before you publish it. You don’t want to accidentally leave anyone out!
Other Acknowledgement Article:
- Acknowledgement For Social Science Project
- Acknowledgement For It Project
- Acknowledgement For Account Project
Acknowledgement Examples For Research Paper
There are many different ways to write an acknowledgement for a research paper, and the best way to do it will vary depending on the project and the circumstances. However, some elements are common to most acknowledgements, such as thankfulness to supervisors, colleagues, and co-authors. Acknowledgements can also be a good way to signal the end of a research project.
Leave a Comment Cancel reply
Save my name, email, and website in this browser for the next time I comment.
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 08 April 2024
Large-scale phenotyping of patients with long COVID post-hospitalization reveals mechanistic subtypes of disease
- Felicity Liew 1 na1 ,
- Claudia Efstathiou ORCID: orcid.org/0000-0001-6125-8126 1 na1 ,
- Sara Fontanella 1 ,
- Matthew Richardson 2 ,
- Ruth Saunders 2 ,
- Dawid Swieboda 1 ,
- Jasmin K. Sidhu 1 ,
- Stephanie Ascough 1 ,
- Shona C. Moore ORCID: orcid.org/0000-0001-8610-2806 3 ,
- Noura Mohamed 4 ,
- Jose Nunag ORCID: orcid.org/0000-0002-4218-0500 5 ,
- Clara King 5 ,
- Olivia C. Leavy 2 , 6 ,
- Omer Elneima 2 ,
- Hamish J. C. McAuley 2 ,
- Aarti Shikotra 7 ,
- Amisha Singapuri ORCID: orcid.org/0009-0002-4711-7516 2 ,
- Marco Sereno ORCID: orcid.org/0000-0003-4573-9303 2 ,
- Victoria C. Harris 2 ,
- Linzy Houchen-Wolloff ORCID: orcid.org/0000-0003-4940-8835 8 ,
- Neil J. Greening ORCID: orcid.org/0000-0003-0453-7529 2 ,
- Nazir I. Lone ORCID: orcid.org/0000-0003-2707-2779 9 ,
- Matthew Thorpe 10 ,
- A. A. Roger Thompson ORCID: orcid.org/0000-0002-0717-4551 11 ,
- Sarah L. Rowland-Jones 11 ,
- Annemarie B. Docherty ORCID: orcid.org/0000-0001-8277-420X 10 ,
- James D. Chalmers 12 ,
- Ling-Pei Ho ORCID: orcid.org/0000-0001-8319-301X 13 ,
- Alexander Horsley ORCID: orcid.org/0000-0003-1828-0058 14 ,
- Betty Raman 15 ,
- Krisnah Poinasamy 16 ,
- Michael Marks 17 , 18 , 19 ,
- Onn Min Kon 1 ,
- Luke S. Howard ORCID: orcid.org/0000-0003-2822-210X 1 ,
- Daniel G. Wootton 3 ,
- Jennifer K. Quint 1 ,
- Thushan I. de Silva ORCID: orcid.org/0000-0002-6498-9212 11 ,
- Antonia Ho 20 ,
- Christopher Chiu ORCID: orcid.org/0000-0003-0914-920X 1 ,
- Ewen M. Harrison ORCID: orcid.org/0000-0002-5018-3066 10 ,
- William Greenhalf 21 ,
- J. Kenneth Baillie ORCID: orcid.org/0000-0001-5258-793X 10 , 22 , 23 ,
- Malcolm G. Semple ORCID: orcid.org/0000-0001-9700-0418 3 , 24 ,
- Lance Turtle 3 , 24 ,
- Rachael A. Evans ORCID: orcid.org/0000-0002-1667-868X 2 ,
- Louise V. Wain 2 , 6 ,
- Christopher Brightling 2 ,
- Ryan S. Thwaites ORCID: orcid.org/0000-0003-3052-2793 1 na1 ,
- Peter J. M. Openshaw ORCID: orcid.org/0000-0002-7220-2555 1 na1 ,
- PHOSP-COVID collaborative group &
ISARIC investigators
Nature Immunology volume 25 , pages 607–621 ( 2024 ) Cite this article
13k Accesses
1 Citations
739 Altmetric
Metrics details
- Inflammasome
- Inflammation
- Innate immunity
One in ten severe acute respiratory syndrome coronavirus 2 infections result in prolonged symptoms termed long coronavirus disease (COVID), yet disease phenotypes and mechanisms are poorly understood 1 . Here we profiled 368 plasma proteins in 657 participants ≥3 months following hospitalization. Of these, 426 had at least one long COVID symptom and 233 had fully recovered. Elevated markers of myeloid inflammation and complement activation were associated with long COVID. IL-1R2, MATN2 and COLEC12 were associated with cardiorespiratory symptoms, fatigue and anxiety/depression; MATN2, CSF3 and C1QA were elevated in gastrointestinal symptoms and C1QA was elevated in cognitive impairment. Additional markers of alterations in nerve tissue repair (SPON-1 and NFASC) were elevated in those with cognitive impairment and SCG3, suggestive of brain–gut axis disturbance, was elevated in gastrointestinal symptoms. Severe acute respiratory syndrome coronavirus 2-specific immunoglobulin G (IgG) was persistently elevated in some individuals with long COVID, but virus was not detected in sputum. Analysis of inflammatory markers in nasal fluids showed no association with symptoms. Our study aimed to understand inflammatory processes that underlie long COVID and was not designed for biomarker discovery. Our findings suggest that specific inflammatory pathways related to tissue damage are implicated in subtypes of long COVID, which might be targeted in future therapeutic trials.
Similar content being viewed by others
Immunopathological signatures in multisystem inflammatory syndrome in children and pediatric COVID-19
Keith Sacco, Riccardo Castagnoli, … Luigi D. Notarangelo
In COVID-19, NLRP3 inflammasome genetic variants are associated with critical disease and these effects are partly mediated by the sickness symptom complex: a nomothetic network approach
Michael Maes, Walton Luiz Del Tedesco Junior, … Andréa Name Colado Simão
Epidemiology, clinical presentation, pathophysiology, and management of long COVID: an update
Sizhen Su, Yimiao Zhao, … Lin Lu
One in ten severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections results in post-acute sequelae of coronavirus disease 2019 (PASC) or long coronavirus disease (COVID), which affects 65 million people worldwide 1 . Long COVID (LC) remains common, even after mild acute infection with recent variants 2 , and it is likely LC will continue to cause substantial long-term ill health, requiring targeted management based on an understanding of how disease phenotypes relate to underlying mechanisms. Persistent inflammation has been reported in adults with LC 1 , 3 , but studies have been limited in size, timing of samples or breadth of immune mediators measured, leading to inconsistent or absent associations with symptoms. Markers of oxidative stress, metabolic disturbance, vasculoproliferative processes and IFN-, NF-κB- or monocyte-related inflammation have been suggested 3 , 4 , 5 , 6 .
The PHOSP-COVID study, a multicenter United Kingdom study of patients previously hospitalized with COVID-19, has reported inflammatory profiles in 626 adults with health impairment after COVID-19, identified through clustering. Elevated IL-6 and markers of mucosal inflammation were observed in those with severe impairment compared with individuals with milder impairment 7 . However, LC is a heterogeneous condition that may be a distinct form of health impairment after COVID-19, and it remains unclear whether there are inflammatory changes specific to LC symptom subtypes. Determining whether activated inflammatory pathways underlie all cases of LC or if mechanisms differ according to clinical presentation is essential for developing effective therapies and has been highlighted as a top research priority by patients and clinicians 8 .
In this Letter, in a prospective multicenter study, we measured 368 plasma proteins in 657 adults previously hospitalized for COVID-19 (Fig. 1a and Table 1 ). Individuals in our cohort experienced a range of acute COVID-19 severities based on World Health Organization (WHO) progression scores 9 ; WHO 3–4 (no oxygen support, n = 133 and median age of 55 years), WHO 5–6 (oxygen support, n = 353 and median age of 59 years) and WHO 7–9 (critical care, n = 171 and median age of 57 years). Participants were hospitalized for COVID-19 ≥3 months before sample collection (median 6.1 months, interquartile range (IQR) 5.1–6.8 months and range 3.0–8.3 months) and confirmed clinically ( n = 36/657) or by PCR ( n = 621/657). Symptom data indicated 233/657 (35%) felt fully recovered at 6 months (hereafter ‘recovered’) and the remaining 424 (65%) reported symptoms consistent with the WHO definition for LC (symptoms ≥3 months post infection 10 ). Given the diversity of LC presentations, patients were grouped according to symptom type (Fig. 1b ). Groups were defined using symptoms and health deficits that have been commonly reported in the literature 1 ( Methods ). A multivariate penalized logistic regression model (PLR) was used to explore associations of clinical covariates and immune mediators at 6 months between recovered patients ( n = 233) and each LC group (cardiorespiratory symptoms, cardioresp, n = 398, Fig. 1c ; fatigue, n = 384, Fig. 1d ; affective symptoms, anxiety/depression, n = 202, Fig. 1e ; gastrointestinal symptoms, GI, n = 132, Fig. 1f ; and cognitive impairment, cognitive, n = 61, Fig. 1g ). Women ( n = 239) were more likely to experience CardioResp (odds ratio (OR 1.14), Fatigue (OR 1.22), GI (OR 1.13) and Cognitive (OR 1.03) outcomes (Fig. 1c,d,f,g ). Repeated cross-validation was used to optimize and assess model performance ( Methods and Extended Data Fig. 1 ). Pre-existing conditions, such as chronic lung disease, neurological disease and cardiovascular disease (Supplementary Table 1 ), were associated with all LC groups (Fig. 1c–g ). Age, C-reactive protein (CRP) and acute disease severity were not associated with any LC group (Table 1 ).
a , Distribution of time from COVID-19 hospitalization at sample collection. All samples were cross-sectional. The vertical red line indicates the 3 month cutoff used to define our final cohort and samples collected before 3 months were excluded. b , An UpSet plot describing pooled LC groups. The horizontal colored bars represent the number of patients in each symptom group: cardiorespiratory (Cardio_Resp), fatigue, cognitive, GI and anxiety/depression (Anx_Dep). Vertical black bars represent the number of patients in each symptom combination group. To prevent patient identification, where less than five patients belong to a combination group, this has been represented as ‘<5’. The recovered group ( n = 233) were used as controls. c – g , Forest plots of Olink protein concentrations (NPX) associated with Cardio_Resp ( n = 365) ( c ), fatigue (n = 314) ( d ), Anx_Dep ( n = 202) ( e ), GI ( n = 124) ( f ) and cognitive ( n = 60) ( g ). Neuro_Psych, neuropsychiatric. The error bars represent the median accuracy of the model. h , i , Distribution of Olink values (NPX) for IL-1R2 ( h ) and MATN2, neurofascin and sCD58 ( i ) measured between symptomatic and recovered individuals in recovered ( n = 233), Cardio_Resp ( n = 365), fatigue ( n = 314) and Anx_Dep ( n = 202) groups ( h ) and MATN2 in GI ( n = 124), neurofascin in cognitive ( n = 60) and sCD58 in Cardio_Resp and recovered groups ( i ). The box plot center line represents the median, the boundaries represent IQR and the whisker length represents 1.5× IQR. The median values were compared between groups using two-sided Wilcoxon signed-rank test, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001.
To study the association of peripheral inflammation with symptoms, we analyzed cross-sectional data collected approximately 6 months after hospitalizations. We measured 368 immune mediators from plasma collected contemporaneously with symptom data. Mediators suggestive of myeloid inflammation were associated with all symptoms (Fig. 1c–h ). Elevated IL-1R2, an IL-1 receptor expressed by monocytes and macrophages modulating inflammation 11 and MATN2, an extracellular matrix protein that modulates tissue inflammation through recruitment of innate immune cells 12 , were associated with cardioresp (IL-1R2 OR 1.14, Fig. 1c,h ), fatigue (IL-1R2 OR 1.45, Fig. 1d,h ), anxiety/depression (IL-1R2 OR 1.34. Fig. 1e,h ) and GI (MATN2 OR 1.08, Fig. 1f ). IL-3RA, an IL-3 receptor, was associated with cardioresp (OR 1.07, Fig. 1c ), fatigue (OR 1.21, Fig. 1d ), anxiety/depression (OR 1.12, Fig. 1e ) and GI (OR 1.06, Fig. 1f ) groups, while CSF3, a cytokine promoting neutrophilic inflammation 13 , was elevated in cardioresp (OR 1.06, Fig. 1c ), fatigue (OR 1.12, Fig. 1d ) and GI (OR 1.08, Fig. 1f ).
Elevated COLEC12, which initiates inflammation in tissues by activating the alternative complement pathway 14 , associated with cardioresp (OR 1.09, Fig. 1c ), fatigue (OR 1.19, Fig. 1d ) and anxiety/depression (OR 1.11, Fig. 1e ), but not with GI (Fig. 1f ) and only weakly with cognitive (OR 1.02, Fig. 1g ). C1QA, a degradation product released by complement activation 15 was associated with GI (OR 1.08, Fig. 1f ) and cognitive (OR 1.03, Fig. 1g ). C1QA, which is known to mediate dementia-related neuroinflammation 16 , had the third strongest association with cognitive (Fig. 1g ). These observations indicated that myeloid inflammation and complement activation were associated with LC.
Increased expression of DPP10 and SCG3 was observed in the GI group compared with recovered (DPP10 OR 1.07 and SCG3 OR 1.08, Fig. 1f ). DPP10 is a membrane protein that modulates tissue inflammation, and increased DPP10 expression is associated with inflammatory bowel disease 17 , 18 , suggesting that GI symptoms may result from enteric inflammation. Elevated SCG3, a multifunctional protein that has been associated with irritable bowel syndrome 19 , suggested that noninflammatory disturbance of the brain–gut axis or dysbiosis, may occur in the GI group. The cognitive group was associated with elevated CTSO (OR 1.04), NFASC (OR 1.03) and SPON-1 (OR 1.02, Fig. 1g,i ). NFASC and SPON-1 regulate neural growth 20 , 21 , while CTSO is a cysteine proteinase supporting tissue turnover 22 . The increased expression of these three proteins as well as C1QA and DPP10 in the cognitive group (Fig. 1g ) suggested neuroinflammation and alterations in nerve tissue repair, possibly resulting in neurodegeneration. Together, our findings indicated that complement activation and myeloid inflammation were common to all LC groups, but subtle differences were observed in the GI and cognitive groups, which may have mechanistic importance. Acutely elevated fibrinogen during hospitalization has been reported to be predictive of LC cognitive deficits 23 . We found elevated fibrinogen in LC relative to recovered (Extended Data Fig. 2a ; P = 0.0077), although this was not significant when restricted to the cognitive group ( P = 0.074), supporting our observation of complement pathway activation in LC and in keeping with reports that complement dysregulation and thrombosis drive severe COVID-19 (ref. 24 ).
Elevated sCD58 was associated with lower odds of all LC symptoms and was most pronounced in cardioresp (OR 0.85, Fig. 1c,i ), fatigue (OR 0.80, Fig. 1d ) and anxiety/depression (OR 0.83, Fig. 1e ). IL-2 was negatively associated with the cardioresp (Fig. 1c , OR 0.87), fatigue (Fig. 1d , OR 0.80), anxiety/depression (Fig. 1e , OR 0.84) and cognitive (Fig. 1g , OR 0.96) groups. Both IL-2 and sCD58 have immunoregulatory functions 25 , 26 . Specifically, sCD58 suppresses IL-1- or IL-6-dependent interactions between CD2 + monocytes and CD58 + T or natural killer cells 26 . The association of sCD58 with recovered suggests a central role of dysregulated myeloid inflammation in LC. Elevated markers of tissue repair, IDS and DNER 27 , 28 , were also associated with recovered relative to all LC groups (Fig. 1c–g ). Taken together, our data suggest that suppression of myeloid inflammation and enhanced tissue repair were associated with recovered, supporting the use of immunomodulatory agents in therapeutic trials 29 (Supplementary Table 2 ).
We next sought to validate the experimental and analytical approaches used. Although Olink has been validated against other immunoassay platforms, showing superior sensitivity and specificity 30 , 31 , we confirmed the performance of Olink against chemiluminescent immunoassays within our cohort. We performed chemiluminescent immunoassays on plasma from a subgroup of 58 participants (recovered n = 13 and LC n = 45). There were good correlations between results from Olink (normalized protein expression (NPX)) and chemiluminescent immunoassays (pg ml −1 ) for CSF3, IL-1R2, IL-3RA, TNF and TFF2 (Extended Data Fig. 3 ). Most samples did not have concentrations of IL-2 detectable using a mesoscale discovery chemiluminescent assay, limiting this analysis to 14 samples (recovered n = 4, LC n = 10, R = 0.55 and P = 0.053, Extended Data Fig. 3 ). We next repeated our analysis using alternative definitions of LC. The Centers for Disease Control and Prevention and National Institute for Health and Care Excellence definitions for LC include symptoms occurring 1 month post infection 32 , 33 . Using the 1 month post-infection definition included 62 additional participants to our analysis (recovered n = 21, 3 females and median age 61 years and LC n = 41, 15 females and median age 60 years, Extended Data Fig. 2c ) and found that inflammatory associations with each LC group were consistent with our analysis based on the WHO definition (Extended Data Fig. 2d–h ). Finally, to validate the analytical approach (PLR) we examined the distribution of data, prioritizing proteins that were most strongly associated with each LC/recovered group (IL-1R2, MATN2, NFASC and sCD58). Each protein was significantly elevated in the LC group compared with recovered (Fig. 1h,i and Extended Data Fig. 4 ), consistent with the PLR. Alternative regression approaches (unadjusted regression models and partial least squares, PLS) reported results consistent with the original analysis of protein associations and LC outcome in the WHO-defined cohort (Fig. 1c–g , Supplementary Table 3 and Extended Data Figs. 5 and 6 ). The standard errors of PLS estimates were wide (Extended Data Fig. 6 ), consistent with previous demonstrations that PLR is the optimal method to analyze high-dimensional data where variables may have combined effects 34 . As inflammatory proteins are often colinear, working in-tandem to mediate effects, we prioritized PLR results to draw conclusions.
To explore the relationship between inflammatory mediators associated with different LC symptoms, we performed a network analysis of Olink mediators highlighted by PLR within each LC group. COLEC12 and markers of endothelial and mucosal inflammation (MATN2, PCDH1, ROBO1, ISM1, ANGPTL2, TGF-α and TFF2) were highly correlated within the cardioresp, fatigue and anxiety/depression groups (Fig. 2 and Extended Data Fig. 7 ). Elevated PCDH1, an adhesion protein modulating airway inflammation 35 , was highly correlated with other inflammatory proteins associated with the cardioresp group (Fig. 2 ), suggesting that systemic inflammation may arise from the lung in these individuals. This was supported by increased expression of IL-3RA, which regulates innate immune responses in the lung through interactions with circulating IL-3 (ref. 36 ), in fatigue (Figs. 1d and 2 ), which correlated with markers of tissue inflammation, including PCDH1 (Fig. 2 ). MATN2 and ISM1, mucosal proteins that enhance inflammation 37 , 38 , were highly correlated in the GI group (Fig. 2 ), highlighting the role of tissue-specific inflammation in different LC groups. SCG3 correlated less closely with mediators in the GI group (Fig. 2 ), suggesting that the brain–gut axis may contribute separately to some GI symptoms. SPON-1, which regulates neural growth 21 , was the most highly correlated mediator in the cognitive group (Fig. 2 and Extended Data Fig. 7 ), highlighting that processes within nerve tissue may underlie this group. These observations suggested that inflammation might arise from mucosal tissues and that additional mechanisms may contribute to pathophysiology underlying the GI and cognitive groups.
Network analysis of Olink mediators associated with cardioresp ( n = 365), fatigue ( n = 314), anxiety/depression ( n = 202), GI ( n = 124) and cognitive groups ( n = 60). Each node corresponds to a protein mediator identified by PLR. The edges (blue lines) were weighted according to the size of Spearman’s rank correlation coefficient between proteins. All edges represent positive and significant correlations ( P < 0.05) after FDR adjustment.
Women were more likely to experience LC (Table 1 ), as found in previous studies 1 . As estrogen can influence immunological responses 39 , we investigated whether hormonal differences between men and women with LC in our cohort explained this trend. We grouped men and women with LC symptoms into two age groups (those younger than 50 years and those 50 years and older, using age as a proxy for menopause status in women) and compared mediator levels between men and women in each age group, prioritizing those identified by PLR to be higher in LC compared with recovered. As we aimed to understand whether women with LC had stronger inflammatory responses than men with LC, we did not assess differences in men and women in the recovered group. IL-1R2 and MATN2 were significantly higher in women ≥50 years than men ≥50 years in the cardioresp group (Fig. 3a , IL-1R2 and MATN2) and the fatigue group (Fig. 3b ). In the GI group, CSF3 was higher in women ≥50 years compared with men ≥50 years (Fig. 3c ), indicating that the inflammatory markers observed in women were not likely to be estrogen-dependent. Women have been reported to have stronger innate immune responses to infection and to be at greater risk of autoimmunity 39 , possibly explaining why some women in the ≥50 years group had higher inflammatory proteins than men the same group. Proteins associated with the anxiety/depression (IL-1R2 P = 0.11 and MATN2 P = 0.61, Extended Data Fig. 8a ) and cognitive groups (CTSO P = 0.64 and NFASC P = 0.41, Extended Data Fig. 8b ) were not different between men and women in either age group, consistent with the absent/weak association between sex and these outcomes identified by PLR (Fig. 1e,g ). Though our findings suggested that nonhormonal differences in inflammatory responses may explain why some women are more likely to have LC, they require confirmation in adequately powered studies.
a – c , Olink-measured plasma protein levels (NPX) of IL-1R2 and MATN2 ( a and b ) and CSF3 ( c ) between LC men and LC women divided by age (<50 or ≥50 years) in the cardiorespiratory group (<50 years n = 8 and ≥50 years n = 270) ( a ), fatigue group (<50 years n = 81 and ≥50 years n = 227) ( b ) and GI group (<50 years n = 34 and ≥50 years n = 82) ( c ). the median values were compared between men and women using two-sided Wilcoxon signed-rank test, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001. The box plot center line represents the median, the boundaries represent IQR and the whisker length represents 1.5× IQR.
To test whether local respiratory tract inflammation persisted after COVID-19, we compared nasosorption samples from 89 participants (recovered, n = 31; LC, n = 33; and healthy SARS-CoV-2 naive controls, n = 25, Supplementary Tables 4 and 5 ). Several inflammatory markers were elevated in the upper respiratory tract post COVID (including IL-1α, CXCL10, CXCL11, TNF, VEGF and TFF2) when compared with naive controls, but similar between recovered and LC (Fig. 4a ). In the cardioresp group ( n = 29), inflammatory mediators elevated in plasma (for example, IL-6, APO-2, TGF-α and TFF2) were not elevated in the upper respiratory tract (Extended Data Fig. 9a ) and there was no correlation between plasma and nasal mediator levels (Extended Data Fig. 9b ). This exploratory analysis suggested upper respiratory tract inflammation post COVID was not specifically associated with cardiorespiratory symptoms.
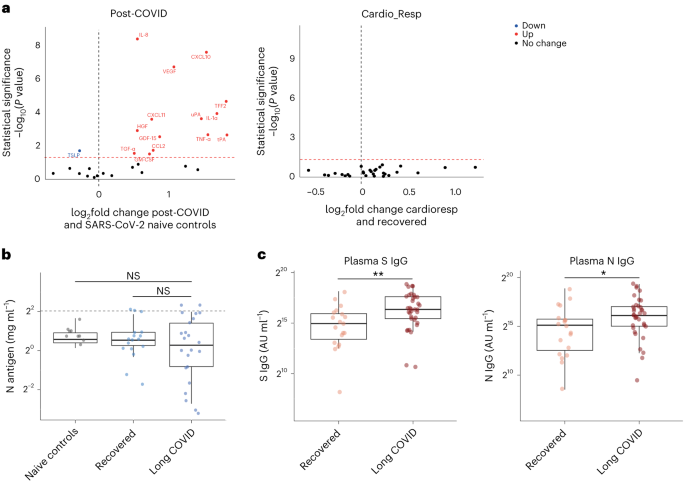
a , Nasal cytokines measured by immunoassay in post-COVID participants ( n = 64) compared with healthy SARS-CoV-2 naive controls ( n = 25), and between the the cardioresp group ( n = 29) and the recovered group ( n = 31). The red values indicate significantly increased cytokine levels after FDR adjustment ( P < 0.05) using two-tailed Wilcoxon signed-rank test. b , SARS-CoV-2 N antigen measured in sputum by electrochemiluminescence from recovered ( n = 17) and pooled LC ( n = 23) groups, compared with BALF from SARS-CoV-2 naive controls ( n = 9). The horizontal dashed line indicates the lower limit of detection of the assay. c , Plasma S- and N-specific IgG responses measured by electrochemiluminescence in the LC ( n = 35) and recovered ( n = 19) groups. The median values were compared using two-sided Wilcoxon signed-rank tests, NS P > 0.05, * P < 0.05, ** P < 0.01, *** P < 0.001 and **** P < 0.0001. The box plot center lines represent the median, the boundaries represent IQR and the whisker length represents 1.5× IQR.
To explore whether SARS-CoV-2 persistence might explain the inflammatory profiles observed in the cardioresp group, we measured SARS-CoV-2 nucleocapsid (N) antigen in sputum from 40 participants (recovered n = 17 and LC n = 23) collected approximately 6 months post hospitalization (Supplementary Table 6 ). All samples were compared with prepandemic bronchoalveolar lavage fluid ( n = 9, Supplementary Table 4 ). Only four samples (recovered n = 2 and LC n = 2) had N antigen above the assay’s lower limit of detection, and there was no difference in N antigen concentrations between LC and recovered (Fig. 4b , P = 0.78). These observations did not exclude viral persistence, which might require tissues samples for detection 40 , 41 . On the basis of the hypothesis that persistent viral antigen might prevent a decline in antibody levels over time, we examined the titers of SARS-CoV-2-specific antibodies in unvaccinated individuals (recovered n = 19 and LC n = 35). SARS-CoV-2 N-specific ( P = 0.023) and spike (S)-specific ( P = 0.0040) immunoglobulin G (IgG) levels were elevated in LC compared with recovered (Fig. 4c ).
Overall, we identified myeloid inflammation and complement activation in the cardioresp, fatigue, anxiety/depression, cognitive and GI groups 6 months after hospitalization (Extended Data Fig. 10 ). Our findings build on results of smaller studies 5 , 6 , 42 and are consistent with a genome-wide association study that identified an independent association between LC and FOXP4 , which modulates neutrophilic inflammation and immune cell function 43 , 44 . In addition, we identified tissue-specific inflammatory elements, indicating that myeloid disturbance in different tissues may result in distinct symptoms. Multiple mechanisms for LC have been suggested, including autoimmunity, thrombosis, vascular dysfunction, SARS-CoV-2 persistence and latent virus reactivation 1 . All these processes involve myeloid inflammation and complement activation 45 . Complement activation in LC has been suggested in a proteomic study in 97 mostly nonhospitalized COVID-19 cases 42 and a study of 48 LC patients, of which one-third experienced severe acute disease 46 . As components of the complement system are known to have a short half-life 47 , ongoing complement activation suggests active inflammation rather than past tissue damage from acute infection.
Despite the heterogeneity of LC and the likelihood of coexisting or multiple etiologies, our work suggests some common pathways that might be targeted therapeutically and supports the rationale for several drugs currently under trial. Our finding of increased sCD58 levels (associated with suppression of monocyte–lymphocyte interactions 26 ) in the recovered group, strengthens our conclusion that myeloid inflammation is central to the biology of LC and that trials of steroids, IL-1 antagonists, JAK inhibitors, naltrexone and colchicine are justified. Although anticoagulants such as apixaban might prevent thrombosis downstream of complement dysregulation, they can also increase the risk of serious bleeding when given after COVID-19 hospitalization 48 . Thus, clinical trials, already underway, need to carefully assess the risks and benefits of anticoagulants (Supplementary Table 2 ).
Our finding of elevated S- and N-specific IgG in LC could suggest viral persistence, as found in other studies 6 , 42 , 49 . Our network analysis indicated that inflammatory proteins in the cardioresp group interacted strongly with ISM1 and ROBO1, which are expressed during respiratory tract infection and regulate lung inflammation 50 , 51 . Although we were unable to find SARS-CoV-2 antigen in sputum from our LC cases, we did not test for viral persistence in GI tract and lung tissue 40 , 41 or in plasma 52 . Evidence of SARS-CoV-2 persistence would justify trials of antiviral drugs (singly or in combination) in LC. It is also possible that autoimmune processes could result in an innate inflammatory profile in LC. Autoreactive B cells have been identified in LC patients with higher SARS-CoV-2-specific antibody titers in a study of mostly mild acute COVID cases (59% WHO 2–3) 42 , a different population from our study of hospitalized cases.
Our observations of distinct protein profiles in GI and cognitive groups support previous reports on distinct associations between Epstein–Barr virus reactivation and neurological symptoms, or autoantibodies and GI symptoms relative to other forms of LC 49 , 53 . We did not assess autoantibody induction but found evidence of brain–gut axis disturbance (SCG3) in the GI group, which occurs in many autoimmune diseases 54 . We found signatures suggestive of neuroinflammation (C1QA) in the cognitive group, consistent with findings of brain abnormalities on magnetic resonance imaging after COVID-19 hospitalization 55 , as well as findings of microglial activation in mice after COVID-19 (ref. 56 ). Proinflammatory signatures dominated in the cardioresp, fatigue and anxiety/depression groups and were consistent with those seen in non-COVID depression, suggesting shared mechanisms 57 . The association between markers of myeloid inflammation, including IL-3RA, and symptoms was greatest for fatigue. Whilst membrane-bound IL-3RA facilitates IL-3 signaling upstream of myelopoesis 36 its soluble form (measured in plasma) can bind IL-3 and can act as a decoy receptor, preventing monocyte maturation and enhancing immunopathology 58 . Monocytes from individuals with post-COVID fatigue are reported to have abnormal expression profiles (including reduced CXCR2), suggestive of altered maturation and migration 5 , 59 . Lung-specific inflammation was suggested by the association between PCDH1 (an airway epithelial adhesion molecule 35 ) and cardioresp symptoms.
Our observations do not align with all published observations on LC. One proteomic study of 55 LC cases after generally mild (WHO 2–3) acute disease found that TNF and IFN signatures were elevated in LC 3 . Vasculoproliferative processes and metabolic disturbance have been reported in LC 4 , 60 , but these studies used uninfected healthy individuals for comparison and cannot distinguish between LC-specific phenomena and residual post-COVID inflammation. A study of 63 adults (LC, n = 50 and recovered, n = 13) reported no association between immune cell activation and LC 3 months after infection 61 , though myeloid inflammation was not directly measured, and 3 months post infection may be too early to detect subtle differences between LC and recovered cases due to residual acute inflammation.
Our study has limitations. We designed the study to identify inflammatory markers identifying pathways underlying LC subgroups rather than diagnostic biomarkers. The ORs we report are small, but associations were consistent across alternative methods of analysis and when using different LC definitions. Small effect sizes can be expected when using PLR, which shrinks correlated mediator coefficients to reflect combined effects and prevent colinear inflation 62 , and could also result from measurement of plasma mediators that may underestimate tissue inflammation. Although our LC cohort is large compared with most other published studies, some of our subgroups are small (only 60 cases were designated cognitive). Though the performance of the cognitive PLR model was adequate, our findings should be validated in larger studies. It should be noted that our cohort of hospitalized cases may not represent all types of LC, especially those occurring after mild infection. We looked for an effect of acute disease severity within our study and did not find it, and are reassured that the inflammatory profiles we observed were consistent with those seen in smaller studies including nonhospitalized cases 42 , 46 . Studies of posthospital LC may be confounded by ‘posthospital syndrome’, which encompasses general and nonspecific effects of hospitalization (particularly intensive care) 63 .
In conclusion, we found markers of myeloid inflammation and complement activation in our large prospective posthospital cohort of patients with LC, in addition to distinct inflammatory patterns in patients with cognitive impairment or gastrointestinal symptoms. These findings show the need to consider subphenotypes in managing patients with LC and support the use of antiviral or immunomodulatory agents in controlled therapeutic trials.
Study design and ethics
After hospitalization for COVID-19, adults who had no comorbidity resulting in a prognosis of less than 6 months were recruited to the PHOSP-COVID study ( n = 719). Patients hospitalized between February 2020 and January 2021 were recruited. Both sexes were recruited and gender was self-reported (female, n = 257 and male, n = 462). Written informed consent was obtained from all patients. Ethical approvals for the PHOSP-COVID study were given by Leeds West Research Ethics Committee (20/YH/0225).
Symptom data and samples were prospectively collected from individuals approximately 6 months (IQR 5.1–6.8 months and range 3.0–8.3 months) post hospitalization (Fig. 1a ), via the PHOSP-COVID multicenter United Kingdom study 64 . Data relating to patient demographics and acute admission were collected via the International Severe Acute Respiratory and Emerging Infection Consortium World Health Organization Clinical Characterisation Protocol United Kingdom (ISARIC4C study; IRAS260007/IRAS126600) (ref. 65 ). Adults hospitalized during the SARS-CoV-2 pandemic were systematically recruited into ISARIC4C. Written informed consent was obtained from all patients. Ethical approval was given by the South Central–Oxford C Research Ethics Committee in England (reference 13:/SC/0149), Scotland A Research Ethics Committee (20/SS/0028) and WHO Ethics Review Committee (RPC571 and RPC572l, 25 April 2013).
Data were collected to account for variables affecting symptom outcome, via hospital records and self-reporting. Acute disease severity was classified according to the WHO clinical progression score: WHO class 3–4: no oxygen therapy; class 5: oxygen therapy; class 6: noninvasive ventilation or high-flow nasal oxygen; and class 7–9: managed in critical care 9 . Clinical data were used to place patients into six categories: ‘recovered’, ‘GI’, ‘cardiorespiratory’, ‘fatigue’, ‘cognitive impairment’ and ‘anxiety/depression’ (Supplementary Table 7 ). Patient-reported symptoms and validated clinical scores were used when feasible, including Medical Research Council (MRC) breathlessness score, dyspnea-12 score, Functional Assessment of Chronic Illness Therapy (FACIT) score, Patient Health Questionnaire (PHQ)-9 and Generalized Anxiety Disorder (GAD)-7. Cognitive impairment was defined as a Montreal Cognitive Assessment score <26. GI symptoms were defined as answering ‘Yes’ to the presence of at least two of the listed symptoms. ‘Recovered’ was defined by self-reporting. Patients were placed in multiple groups if they experienced a combination of symptoms.
Matched nasal fluid and sputum samples were prospectively collected from a subgroup of convalescent patients approximately 6 months after hospitalization via the PHOSP-COVID study. Nasal and bronchoalveolar lavage fluid (BALF) collected from healthy volunteers before the COVID-19 pandemic were used as controls (Supplementary Table 4 ). Written consent was obtained for all individuals and ethical approvals were given by London–Harrow Research Ethics Committee (13/LO/1899) for the collection of nasal samples and the Health Research Authority London–Fulham Research Ethics Committee (IRAS project ID 154109; references 14/LO/1023, 10/H0711/94 and 11/LO/1826) for BALF samples.
Ethylenediaminetetraacetic acid plasma was collected from whole blood taken by venepuncture and frozen at −80 °C as previously described 7 , 66 . Nasal fluid was collected using a NasosorptionTM FX·I device (Hunt Developments), which uses a synthetic absorptive matrix to collect concentrated nasal fluid. Samples were eluted and stored as previously described 67 . Sputum samples were collected via passive expectoration and frozen at −80 °C without the addition of buffers. Sputum samples from convalescent individuals were compared with BALF from healthy SARS-CoV-2-naive controls, collected before the pandemic. BALF samples were used to act as a comparison for lower respiratory tract samples since passively expectorated sputum from healthy SARS-CoV-2-naive individuals was not available. BALF samples were obtained by instillation and recovery of up to 240 ml of normal saline via a fiberoptic bronchoscope. BALF was filtered through 100 µM strainers into sterile 50 ml Falcon tubes, then centrifuged for 10 min at 400 g at 4 °C. The resulting supernatant was transferred into sterile 50 ml Falcon tubes and frozen at −80 °C until use. The full methods for BALF collection and processing have been described previously 68 , 69 .
Immunoassays
To determine inflammatory signatures that associated with symptom outcomes, plasma samples were analyzed on an Olink Explore 384 Inflammation panel 70 . Supplementary Table 8 (Appendix 1 ) lists all the analytes measured. To ensure the validity of results, samples were run in a single batch with the use of negative controls, plate controls in triplicate and repeated measurement of patient samples between plates in duplicate. Samples were randomized between plates according to site and sample collection date. Randomization between plates was blind to LC/recovered outcome. Data were first normalized to an internal extension control that was included in each sample well. Plates were standardized by normalizing to interplate controls, run in triplicate on each plate. Each plate contained a minimum of four patient samples, which were duplicates on another plate; these duplicate pairs allowed any plate to be linked to any other through the duplicates. Data were then intensity normalized across all cohort samples. Finally, Olink results underwent quality control processing and samples or analytes that did not reach quality control standards were excluded. Final normalized relative protein quantities were reported as log 2 NPX values.
To further validate our findings, we performed conventional electrochemiluminescence (ECL) assays and enzyme-linked immunosorbent assay for Olink mediators that were associated with symptom outcome ( Supplementary Methods ). Contemporaneously collected plasma samples were available from 58 individuals. Like most omics platforms, Olink measures relative quantities, so perfect agreement with conventional assays that measure absolute concentrations is not expected.
Sputum samples were thawed before analysis and sputum plugs were extracted with the addition of 0.1% dithiothreitol creating a one in two sample dilution, as previously described 71 . SARS-CoV-2 S and N proteins were measured by ECL S-plex assay at a fixed dilution of one in two (Mesoscale Diagnostics), as per the manufacturers protocol 72 . Control BALF samples were thawed and measured on the same plate, neat. The S-plex assay is highly sensitive in detecting viral antigen in respiratory tract samples 73 .
Nasal cytokines were measured by ECL (mesoscale discovery) and Luminex bead multiplex assays (Biotechne). The full methods and list of analytes are detailed in Supplementary Methods .
Statistics and reproducibility
Clinical data was collected via the PHOSP REDCap database, to which access is available under reasonable request as per the data sharing statement in the manuscript. All analyses were performed within the Outbreak Data Analysis Platform (ODAP). All data and code can be accessed using information in the ‘Data sharing’ and ‘Code sharing’ statements at the end of the manuscript. No statistical method was used to predetermine sample size. Data distribution was assumed to be normal but this was not formally tested. Olink assays and immunoassays were randomized and investigators were blinded to outcomes.
To determine protein signatures that associated with each symptom outcome, a ridge PLR was used. PLR shrinks coefficients to account for combined effects within high-dimensional data, preventing false discovery while managing multicollinearity 34 . Thus, PLR was chosen a priori as the most appropriate model to assess associations between a large number of explanatory variables (that may work together to mediate effects) and symptom outcome 34 , 62 , 70 , 74 . In keeping with our aim to perform an unbiased exploration of inflammatory process, the model alpha was set to zero, facilitating regularization without complete penalization of any mediator. This enabled review of all possible mediators that might associate with LC 62 .
A 50 repeats tenfold nested cross-validation was used to select the optimal lambda for each model and assess its accuracy (Extended Data Fig. 1 ). The performance of the cognitive impairment model was influenced by the imbalance in size of the symptom group ( n = 60) relative to recovered ( n = 250). The model was weighted to account for this imbalance resulting in a sensitivity of 0.98, indicating its validity. We have expanded on the model performance and validation approaches in Supplementary Information .
Age, sex, acute disease severity and preexisting comorbidities were included as covariates in the PLR analysis (Supplementary Tables 1 and 3 ). Covariates were selected a priori using features reported to influence the risk of LC and inflammatory responses 1 , 39 , 64 , 75 . Ethnicity was not included since it has been shown not to predict symptom outcome in this cohort 64 . Individuals with missing data were excluded from the regression analysis. Each symptom group was compared with the ‘recovered’ group. The model coefficients of each covariate were converted into ORs for each outcome and visualized in a forest plot, after removing variables associated with regularized OR between 0.98 and 1.02 or in cases where most variables fell outside of this range, using mediators associated with the highest decile of coefficients either side of this range. This enabled exclusion of mediators with effect sizes that were unlikely to have clinical or mechanistic importance since the ridge PLR shrinks and orders coefficients according to their relative importance rather than making estimates with standard error. Thus, confidence intervals cannot be appropriately derived from PLR, and forest plot error bars were calculated using the median accuracy of the model generated by the nested cross-validation. To verify observations made through PLR analysis, we also performed an unadjusted PLR, an unadjusted logistic regression and a PLS analysis. Univariate analyses using Wilcoxon signed-rank test was also performed (Supplementary Table 8 , Appendix 1 ). Analyses were performed in R version 4.2.0 using ‘data.table v1.14.2’, ‘EnvStats v2.7.0’ ‘tidyverse v1.3.2’, ‘lme4 v1.1-32’, ‘caret v6.0-93’, ‘glmnet v4.1-6’, ‘mdatools v0.14.0’, ‘ggpubbr v0.4.0’ and ‘ggplot2 v3.3.6’ packages.
To further investigate the relationship between proteins elevated in each symptom group, we performed a correlation network analysis using Spearman’s rank correlation coefficient and false discovery rate (FDR) thresholding. The mediators visualized in the PLR forest plots, which were associated with cardiorespiratory symptoms, fatigue, anxiety/depression GI symptoms and cognitive impairment were used, respectively. Analyses were performed in R version 4.2.0 using ‘bootnet v1.5.6 ’ and ‘qgraph v1.9.8 ’ packages.
To determine whether differences in protein levels between men and women related to hormonal differences, we divided each symptom group into premenopausal and postmenopausal groups using an age cutoff of 50 years old. Differences between sexes in each group were determined using the Wilcoxon signed-rank test. To understand whether antigen persistence contributed to inflammation in adults with LC, the median viral antigen concentration from sputum/BALF samples and cytokine concentrations from nasal samples were compared using the Wilcoxon signed-rank test. All tests were two-tailed and statistical significance was defined as a P value < 0.05 after adjustment for FDR ( q -value of 0.05). Analyses were performed in R version 4.2.0 using ‘bootnet v1.5.6’ and ‘qgraph v1.9.8’ packages.
Extended Data Fig. 10 was made using Biorender, accessed at www.biorender.com .
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This is an open access article under the CC BY 4.0 license.
The PHOSP-COVID protocol, consent form, definition and derivation of clinical characteristics and outcomes, training materials, regulatory documents, information about requests for data access, and other relevant study materials are available online at ref. 76 . Access to these materials can be granted by contacting [email protected] and [email protected].
The ISARIC4C protocol, data sharing and publication policy are available at https://isaric4c.net . ISARIC4C’s Independent Data and Material Access Committee welcomes applications for access to data and materials ( https://isaric4c.net ).
The datasets used in the study contain extensive clinical information at an individual level that prevent them from being deposited in an public depository due to data protection policies of the study. Study data can only be accessed via the ODAP, a protected research environment. All data used in this study are available within ODAP and accessible under reasonable request. Data access criteria and information about how to request access is available online at ref. 76 . If criteria are met and a request is made, access can be gained by signing the eDRIS user agreement.
Code availability
Code was written within the ODAP, using R v4.2.0 and publicly available packages (‘data.table v1.14.2’, ‘EnvStats v2.7.0’, ‘tidyverse v1.3.2’, ‘lme4 v1.1-32’, ‘caret v6.0-93’, ‘glmnet v4.1-6’, ‘mdatools v0.14.0’, ‘ggpubbr v0.4.0’, ‘ggplot2 v3.3.6’, ‘bootnet v1.5.6’ and ‘qgraph v1.9.8’ packages). No new algorithms or functions were created and code used in-built functions in listed packages available on CRAN. The code used to generate data and to analyze data is publicly available at https://github.com/isaric4c/wiki/wiki/ISARIC ; https://github.com/SurgicalInformatics/cocin_cc and https://github.com/ClaudiaEfstath/PHOSP_Olink_NatImm .
Davis, H. E., McCorkell, L., Vogel, J. M. & Topol, E. J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21 , 133–146 (2023).
Article CAS PubMed PubMed Central Google Scholar
Antonelli, M., Pujol, J. C., Spector, T. D., Ourselin, S. & Steves, C. J. Risk of long COVID associated with delta versus omicron variants of SARS-CoV-2. Lancet 399 , 2263–2264 (2022).
Talla, A. et al. Persistent serum protein signatures define an inflammatory subcategory of long COVID. Nat. Commun. 14 , 3417 (2023).
Captur, G. et al. Plasma proteomic signature predicts who will get persistent symptoms following SARS-CoV-2 infection. EBioMedicine 85 , 104293 (2022).
Scott, N. A. et al. Monocyte migration profiles define disease severity in acute COVID-19 and unique features of long COVID. Eur. Respir. J. https://doi.org/10.1183/13993003.02226-2022 (2023).
Klein, J. et al. Distinguishing features of Long COVID identified through immune profiling. Nature https://doi.org/10.1038/s41586-023-06651-y (2023).
Article PubMed PubMed Central Google Scholar
Evans, R. A. et al. Clinical characteristics with inflammation profiling of long COVID and association with 1-year recovery following hospitalisation in the UK: a prospective observational study. Lancet Respir. Med . 10 , 761–775 (2022).
Article CAS Google Scholar
Houchen-Wolloff, L. et al. Joint patient and clinician priority setting to identify 10 key research questions regarding the long-term sequelae of COVID-19. Thorax 77 , 717–720 (2022).
Article PubMed Google Scholar
Marshall, J. C. et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 20 , e192–e197 (2020).
Post COVID-19 condition (long COVID). World Health Organization https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition#:~:text=Definition,months%20with%20no%20other%20explanation (2022).
Peters, V. A., Joesting, J. J. & Freund, G. G. IL-1 receptor 2 (IL-1R2) and its role in immune regulation. Brain Behav. Immun. 32 , 1–8 (2013).
Article CAS PubMed Google Scholar
Luo, Z. et al. Monocytes augment inflammatory responses in human aortic valve interstitial cells via β2-integrin/ICAM-1-mediated signaling. Inflamm. Res. 71 , 681–694 (2022).
Bendall, L. J. & Bradstock, K. F. G-CSF: from granulopoietic stimulant to bone marrow stem cell mobilizing agent. Cytokine Growth Factor Rev. 25 , 355–367 (2014).
Ma, Y. J. et al. Soluble collectin-12 (CL-12) is a pattern recognition molecule initiating complement activation via the alternative pathway. J. Immunol. 195 , 3365–3373 (2015).
Laursen, N. S. et al. Functional and structural characterization of a potent C1q inhibitor targeting the classical pathway of the complement system. Front. Immunol. 11 , 1504 (2020).
Dejanovic, B. et al. Complement C1q-dependent excitatory and inhibitory synapse elimination by astrocytes and microglia in Alzheimer’s disease mouse models. Nat. Aging 2 , 837–850 (2022).
Xue, G., Hua, L., Zhou, N. & Li, J. Characteristics of immune cell infiltration and associated diagnostic biomarkers in ulcerative colitis: results from bioinformatics analysis. Bioengineered 12 , 252–265 (2021).
He, T. et al. Integrative computational approach identifies immune‐relevant biomarkers in ulcerative colitis. FEBS Open Bio. 12 , 500–515 (2022).
Sundin, J. et al. Fecal chromogranins and secretogranins are linked to the fecal and mucosal intestinal bacterial composition of IBS patients and healthy subjects. Sci. Rep. 8 , 16821 (2018).
Kriebel, M., Wuchter, J., Trinks, S. & Volkmer, H. Neurofascin: a switch between neuronal plasticity and stability. Int. J. Biochem. Cell Biol. 44 , 694–697 (2012).
Woo, W.-M. et al. The C. elegans F-spondin family protein SPON-1 maintains cell adhesion in neural and non-neural tissues. Development 135 , 2747–2756 (2008).
Yadati, T., Houben, T., Bitorina, A. & Shiri-Sverdlov, R. The ins and outs of cathepsins: physiological function and role in disease management. Cells 9 , 1679 (2020).
Taquet, M. et al. Acute blood biomarker profiles predict cognitive deficits 6 and 12 months after COVID-19 hospitalization. Nat. Med. https://doi.org/10.1038/s41591-023-02525-y (2023).
Siggins, M. K. et al. Alternative pathway dysregulation in tissues drives sustained complement activation and predicts outcome across the disease course in COVID‐19. Immunology 168 , 473–492 (2023).
Pol, J. G., Caudana, P., Paillet, J., Piaggio, E. & Kroemer, G. Effects of interleukin-2 in immunostimulation and immunosuppression. J. Exp. Med. 217 , e20191247 (2020).
Zhang, Y., Liu, Q., Yang, S. & Liao, Q. CD58 immunobiology at a glance. Front. Immunol. 12 , 705260 (2021).
Demydchuk, M. et al. Insights into Hunter syndrome from the structure of iduronate-2-sulfatase. Nat. Commun. 8 , 15786 (2017).
Wang, Z. et al. DNER promotes epithelial–mesenchymal transition and prevents chemosensitivity through the Wnt/β-catenin pathway in breast cancer. Cell Death Dis. 11 , 642 (2020).
Bonilla, H. et al. Therapeutic trials for long COVID-19: a call to action from the interventions taskforce of the RECOVER initiative. Front. Immunol. 14 , 1129459 (2023).
Wik, L. et al. Proximity extension assay in combination with next-generation sequencing for high-throughput proteome-wide analysis. Mol. Cell. Proteomics 20 , 100168 (2021).
Measuring protein biomarkers with Olink—technical comparisons and orthogonal validation. Olink Proteomics https://www.olink.com/content/uploads/2021/09/olink-technical-comparisons-and-orthogonal-validation-1118-v2.0.pdf (2021).
COVID-19 rapid guideline: managing the long-term effects of COVID-19. National Institute for Health and Care Excellence (NICE), Scottish Intercollegiate Guidelines Network (SIGN) and Royal College of General Practitioners (RCGP) https://www.nice.org.uk/guidance/ng188/resources/covid19-rapid-guideline-managing-the-longterm-effects-of-covid19-pdf-51035515742 (2022).
Long COVID or post-COVID conditions. Centers for Disease Control and Prevention https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html#:~:text=Long%20COVID%20is%20broadly%20defined,after%20acute%20COVID%2D19%20infection (2023).
Firinguetti, L., Kibria, G. & Araya, R. Study of partial least squares and ridge regression methods. Commun. Stat. Simul. Comput 46 , 6631–6644 (2017).
Article Google Scholar
Mortensen, L. J., Kreiner-Moller, E., Hakonarson, H., Bonnelykke, K. & Bisgaard, H. The PCDH1 gene and asthma in early childhood. Eur. Respir. J. 43 , 792–800 (2014).
Tong, Y. et al. The RNFT2/IL-3Rα axis regulates IL-3 signaling and innate immunity. JCI Insight 5 , e133652 (2020).
Wu, Y. et al. Effect of ISM1 on the immune microenvironment and epithelial-mesenchymal transition in colorectal cancer. Front. Cell Dev. Biol. 9 , 681240 (2021).
Luo, G. G. & Ou, J. J. Oncogenic viruses and cancer. Virol. Sin. 30 , 83–84 (2015).
Klein, S. L. & Flanagan, K. L. Sex differences in immune responses. Nat. Rev. Immunol. 16 , 626–638 (2016).
Gaebler, C. et al. Evolution of antibody immunity to SARS-CoV-2. Nature 591 , 639–644 (2021).
Bussani, R. et al. Persistent SARS‐CoV‐2 infection in patients seemingly recovered from COVID‐19. J. Pathol. 259 , 254–263 (2023).
Woodruff, M. C. et al. Chronic inflammation, neutrophil activity, and autoreactivity splits long COVID. Nat. Commun. 14 , 4201 (2023).
Lammi, V. et al. Genome-wide association study of long COVID. Preprint at medRxiv https://doi.org/10.1101/2023.06.29.23292056 (2023).
Ismailova, A. et al. Identification of a forkhead box protein transcriptional network induced in human neutrophils in response to inflammatory stimuli. Front. Immunol. 14 , 1123344 (2023).
Beurskens, F. J., van Schaarenburg, R. A. & Trouw, L. A. C1q, antibodies and anti-C1q autoantibodies. Mol. Immunol. 68 , 6–13 (2015).
Cervia-Hasler, C. et al. Persistent complement dysregulation with signs of thromboinflammation in active long Covid. Science 383 , eadg7942 (2024).
Morgan, B. P. & Harris, C. L. Complement, a target for therapy in inflammatory and degenerative diseases. Nat. Rev. Drug Discov. 14 , 857–877 (2015).
Toshner, M. R. et al. Apixaban following discharge in hospitalised adults with COVID-19: preliminary results from a multicentre, open-label, randomised controlled platform clinical trial. Preprint at medRxiv , https://doi.org/10.1101/2022.12.07.22283175 (2022).
Su, Y. et al. Multiple early factors anticipate post-acute COVID-19 sequelae. Cell 185 , 881–895.e20 (2022).
Branchfield, K. et al. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351 , 707–710 (2016).
Rivera-Torruco, G. et al. Isthmin 1 identifies a subset of lung hematopoietic stem cells and it is associated with systemic inflammation. J. Immunol. 202 , 118.18 (2019).
Swank, Z. et al. Persistent circulating severe acute respiratory syndrome coronavirus 2 spike is associated with post-acute coronavirus disease 2019 sequelae. Clin. Infect. Dis. 76 , e487–e490 (2023).
Peluso, M. J. et al. Chronic viral coinfections differentially affect the likelihood of developing long COVID. J. Clin. Invest. 133 , e163669 (2023).
Bellocchi, C. et al. The interplay between autonomic nervous system and inflammation across systemic autoimmune diseases. Int. J. Mol. Sci. 23 , 2449 (2022).
Raman, B. et al. Multiorgan MRI findings after hospitalisation with COVID-19 in the UK (C-MORE): a prospective, multicentre, observational cohort study. Lancet Respir. Med 11 , 1003–1019 (2023).
Fernández-Castañeda, A. et al. Mild respiratory COVID can cause multi-lineage neural cell and myelin dysregulation. Cell 185 , 2452–2468.e16 (2022).
Dantzer, R., O’Connor, J. C., Freund, G. G., Johnson, R. W. & Kelley, K. W. From inflammation to sickness and depression: when the immune system subjugates the brain. Nat. Rev. Neurosci. 9 , 46–56 (2008).
Broughton, S. E. et al. Dual mechanism of interleukin-3 receptor blockade by an anti-cancer antibody. Cell Rep. 8 , 410–419 (2014).
Ley, K., Miller, Y. I. & Hedrick, C. C. Monocyte and macrophage dynamics during atherogenesis. Arterioscler. Thromb. Vasc. Biol. 31 , 1506–1516 (2011).
Iosef, C. et al. Plasma proteome of long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function. J. Transl. Med. 21 , 377 (2023).
Santopaolo, M. et al. Prolonged T-cell activation and long COVID symptoms independently associate with severe COVID-19 at 3 months. eLife 12 , e85009 (2023).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 , 1–22 (2010).
Voiriot, G. et al. Chronic critical illness and post-intensive care syndrome: from pathophysiology to clinical challenges. Ann. Intensive Care 12 , 58 (2022).
Evans, R. A. et al. Physical, cognitive, and mental health impacts of COVID-19 after hospitalisation (PHOSP-COVID): a UK multicentre, prospective cohort study. Lancet Respir. Med 9 , 1275–1287 (2021).
Docherty, A. B. et al. Features of 20,133 UK patients in hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: prospective observational cohort study. BMJ https://doi.org/10.1136/bmj.m1985 (2020).
Elneima, O. et al. Cohort profile: post-hospitalisation COVID-19 study (PHOSP-COVID). Preprint at medRxiv https://doi.org/10.1101/2023.05.08.23289442 (2023).
Liew, F. et al. SARS-CoV-2-specific nasal IgA wanes 9 months after hospitalisation with COVID-19 and is not induced by subsequent vaccination. EBioMedicine 87 , 104402 (2023).
Ascough, S. et al. Divergent age-related humoral correlates of protection against respiratory syncytial virus infection in older and young adults: a pilot, controlled, human infection challenge model. Lancet Healthy Longev. 3 , e405–e416 (2022).
Guvenel, A. et al. Epitope-specific airway-resident CD4+ T cell dynamics during experimental human RSV infection. J. Clin. Invest. 130 , 523–538 (2019).
Article PubMed Central Google Scholar
Greenwood, C. J. et al. A comparison of penalised regression methods for informing the selection of predictive markers. PLoS ONE 15 , e0242730 (2020).
Higham, A. et al. Leukotriene B4 levels in sputum from asthma patients. ERJ Open Res. 2 , 00088–02015 (2016).
SARS-CoV-2 spike kit. MSD https://www.mesoscale.com/~/media/files/product%20inserts/s-plex%20sars-cov-2%20spike%20kit%20product%20insert.pdf (2023).
Ren, A. et al. Ultrasensitive assay for saliva-based SARS-CoV-2 antigen detection. Clin. Chem. Lab. Med. 60 , 771–777 (2022).
Breheny, P. & Huang, J. Penalized methods for bi-level variable selection. Stat. Interface 2 , 369–380 (2009).
Thwaites, R. S. et al. Inflammatory profiles across the spectrum of disease reveal a distinct role for GM-CSF in severe COVID-19. Sci. Immunol. 6 , eabg9873 (2021).
Resources. PHOSP-COVID https://phosp.org/resource/ (2022).
Download references
Acknowledgements
This research used data assets made available by ODAP as part of the Data and Connectivity National Core Study, led by Health Data Research UK in partnership with the Office for National Statistics and funded by UK Research and Innovation (grant ref. MC_PC_20058). This work is supported by the following grants: the PHOSP-COVD study is jointly funded by UK Research and Innovation and National Institute of Health and Care Research (NIHR; grant references MR/V027859/1 and COV0319). ISARIC4C is supported by grants from the National Institute for Health and Care Research (award CO-CIN-01) and the MRC (grant MC_PC_19059) Liverpool Experimental Cancer Medicine Centre provided infrastructure support for this research (grant reference C18616/A25153). Other grants that have supported this work include the UK Coronavirus Immunology Consortium (funder reference 1257927), the Imperial Biomedical Research Centre (NIHR Imperial BRC, grant IS-BRC-1215-20013), the Health Protection Research Unit in Respiratory Infections at Imperial College London and NIHR Health Protection Research Unit in Emerging and Zoonotic Infections at University of Liverpool, both in partnership with Public Health England, (NIHR award 200907), Wellcome Trust and Department for International Development (215091/Z/18/Z), Health Data Research UK (grant code 2021.0155), MRC (grant code MC_UU_12014/12) and NIHR Clinical Research Network for providing infrastructure support for this research. We also acknowledge the support of the MRC EMINENT Network (MR/R502121/1), which is cofunded by GSK, the Comprehensive Local Research Networks, the MRC HIC-Vac network (MR/R005982/1) and the RSV Consortium in Europe Horizon 2020 Framework Grant 116019. F.L. is supported by an MRC clinical training fellowship (award MR/W000970/1). C.E. is funded by NIHR (grant P91258-4). L.-P.H. is supported by Oxford NIHR Biomedical Research Centre. A.A.R.T. is supported by a British Heart Foundation (BHF) Intermediate Clinical Fellowship (FS/18/13/33281). S.L.R.-J. receives support from UK Research and Innovation (UKRI), Global Challenges Research Fund (GCRF), Rosetrees Trust, British HIV association (BHIVA), European & Developing Countries Clinical Trials Partnership (EDCTP) and Globvac. J.D.C. has grants from AstraZeneca, Boehringer Ingelheim, GSK, Gilead Sciences, Grifols, Novartis and Insmed. R.A.E. holds a NIHR Clinician Scientist Fellowship (CS-2016-16-020). A. Horsley is currently supported by UK Research and Innovation, NIHR and NIHR Manchester BRC. B.R. receives support from BHF Oxford Centre of Research Excellence, NIHR Oxford BRC and MRC. D.G.W. is supported by an NIHR Advanced Fellowship. A. Ho has received support from MRC and for the Coronavirus Immunology Consortium (MR/V028448/1). L.T. is supported by the US Food and Drug Administration Medical Countermeasures Initiative contract 75F40120C00085 and the National Institute for Health Research Health Protection Research Unit in Emerging and Zoonotic Infections (NIHR200907) at the University of Liverpool in partnership with UK Health Security Agency (UK-HSA), in collaboration with Liverpool School of Tropical Medicine and the University of Oxford. L.V.W. has received support from UKRI, GSK/Asthma and Lung UK and NIHR for this study. M.G.S. has received support from NIHR UK, MRC UK and Health Protection Research Unit in Emerging and Zoonotic Infections, University of Liverpool. J.K.B. is supported by the Wellcome Trust (223164/Z/21/Z) and UKRI (MC_PC_20004, MC_PC_19025, MC_PC_1905, MRNO2995X/1 and MC_PC_20029). The funders were not involved in the study design, interpretation of data or writing of this manuscript. The views expressed are those of the authors and not necessarily those of the Department of Health and Social Care (DHSC), the Department for International Development (DID), NIHR, MRC, the Wellcome Trust, UK-HSA, the National Health Service or the Department of Health. P.J.M.O. is supported by a NIHR Senior Investigator Award (award 201385). We thank all the participants and their families. We thank the many research administrators, health-care and social-care professionals who contributed to setting up and delivering the PHOSP-COVID study at all of the 65 NHS trusts/health boards and 25 research institutions across the United Kingdom, as well as those who contributed to setting up and delivering the ISARIC4C study at 305 NHS trusts/health boards. We also thank all the supporting staff at the NIHR Clinical Research Network, Health Research Authority, Research Ethics Committee, Department of Health and Social Care, Public Health Scotland and Public Health England. We thank K. Holmes at the NIHR Office for Clinical Research Infrastructure for her support in coordinating the charities group. The PHOSP-COVID industry framework was formed to provide advice and support in commercial discussions, and we thank the Association of the British Pharmaceutical Industry as well the NIHR Office for Clinical Research Infrastructure for coordinating this. We are very grateful to all the charities that have provided insight to the study: Action Pulmonary Fibrosis, Alzheimer’s Research UK, Asthma and Lung UK, British Heart Foundation, Diabetes UK, Cystic Fibrosis Trust, Kidney Research UK, MQ Mental Health, Muscular Dystrophy UK, Stroke Association Blood Cancer UK, McPin Foundations and Versus Arthritis. We thank the NIHR Leicester Biomedical Research Centre patient and public involvement group and Long Covid Support. We also thank G. Khandaker and D. C. Newcomb who provided valuable feedback on this work. Extended Data Fig. 10 was created using Biorender.
Author information
These authors contributed equally: Felicity Liew, Claudia Efstathiou, Ryan S. Thwaites, Peter J. M. Openshaw.
Authors and Affiliations
National Heart and Lung Institute, Imperial College London, London, UK
Felicity Liew, Claudia Efstathiou, Sara Fontanella, Dawid Swieboda, Jasmin K. Sidhu, Stephanie Ascough, Onn Min Kon, Luke S. Howard, Jennifer K. Quint, Christopher Chiu, Ryan S. Thwaites, Peter J. M. Openshaw, Jake Dunning & Peter J. M. Openshaw
Institute for Lung Health, Leicester NIHR Biomedical Research Centre, University of Leicester, Leicester, UK
Matthew Richardson, Ruth Saunders, Olivia C. Leavy, Omer Elneima, Hamish J. C. McAuley, Amisha Singapuri, Marco Sereno, Victoria C. Harris, Neil J. Greening, Rachael A. Evans, Louise V. Wain, Christopher Brightling & Ananga Singapuri
NIHR Health Protection Research Unit in Emerging and Zoonotic Infections, Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK
Shona C. Moore, Daniel G. Wootton, Malcolm G. Semple, Lance Turtle, William A. Paxton & Georgios Pollakis
The Imperial Clinical Respiratory Research Unit, Imperial College NHS Trust, London, UK
Noura Mohamed
Cardiovascular Research Team, Imperial College Healthcare NHS Trust, London, UK
Jose Nunag & Clara King
Department of Population Health Sciences, University of Leicester, Leicester, UK
Olivia C. Leavy, Louise V. Wain & Beatriz Guillen-Guio
NIHR Leicester Biomedical Research Centre, University of Leicester, Leicester, UK
Aarti Shikotra
Centre for Exercise and Rehabilitation Science, NIHR Leicester Biomedical Research Centre-Respiratory, University of Leicester, Leicester, UK
Linzy Houchen-Wolloff
Usher Institute, University of Edinburgh, Edinburgh, UK
Nazir I. Lone, Luke Daines, Annemarie B. Docherty, Nazir I. Lone, Matthew Thorpe, Annemarie B. Docherty, Thomas M. Drake, Cameron J. Fairfield, Ewen M. Harrison, Stephen R. Knight, Kenneth A. Mclean, Derek Murphy, Lisa Norman, Riinu Pius & Catherine A. Shaw
Centre for Medical Informatics, The Usher Institute, University of Edinburgh, Edinburgh, UK
Matthew Thorpe, Annemarie B. Docherty, Ewen M. Harrison, J. Kenneth Baillie, Sarah L. Rowland-Jones, A. A. Roger Thompson & Thushan de Silva
Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, UK
A. A. Roger Thompson, Sarah L. Rowland-Jones, Thushan I. de Silva & James D. Chalmers
University of Dundee, Ninewells Hospital and Medical School, Dundee, UK
James D. Chalmers & Ling-Pei Ho
MRC Human Immunology Unit, University of Oxford, Oxford, UK
Ling-Pei Ho & Alexander Horsley
Division of Infection, Immunity and Respiratory Medicine, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, UK
Alexander Horsley & Betty Raman
Radcliffe Department of Medicine, University of Oxford, Oxford, UK
Betty Raman & Krisnah Poinasamy
Asthma + Lung UK, London, UK
Krisnah Poinasamy & Michael Marks
Department of Clinical Research, London School of Hygiene and Tropical Medicine, London, UK
Michael Marks
Hospital for Tropical Diseases, University College London Hospital, London, UK
Division of Infection and Immunity, University College London, London, UK
Michael Marks & Mahdad Noursadeghi
MRC Centre for Virus Research, School of Infection and Immunity, University of Glasgow, Glasgow, UK
Antonia Ho & William Greenhalf
Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, UK
William Greenhalf & J. Kenneth Baillie
The Roslin Institute, University of Edinburgh, Edinburgh, UK
J. Kenneth Baillie, J. Kenneth Baillie, Sara Clohisey, Fiona Griffiths, Ross Hendry, Andrew Law & Wilna Oosthuyzen
Pandemic Science Hub, University of Edinburgh, Edinburgh, UK
J. Kenneth Baillie
The Pandemic Institute, University of Liverpool, Liverpool, UK
Malcolm G. Semple & Lance Turtle
University of Manchester, Manchester, UK
Kathryn Abel, Perdita Barran, H. Chinoy, Bill Deakin, M. Harvie, C. A. Miller, Stefan Stanel & Drupad Trivedi
Intensive Care Unit, Royal Infirmary of Edinburgh, Edinburgh, UK
Kathryn Abel & J. Kenneth Baillie
North Bristol NHS Trust and University of Bristol, Bristol, UK
H. Adamali, David Arnold, Shaney Barratt, A. Dipper, Sarah Dunn, Nick Maskell, Anna Morley, Leigh Morrison, Louise Stadon, Samuel Waterson & H. Welch
University of Edinburgh, Manchester, UK
Davies Adeloye, D. E. Newby, Riinu Pius, Igor Rudan, Manu Shankar-Hari, Catherine Sudlow, Sarah Walmsley & Bang Zheng
King’s College Hospital NHS Foundation Trust and King’s College London, London, UK
Oluwaseun Adeyemi, Rita Adrego, Hosanna Assefa-Kebede, Jonathon Breeze, S. Byrne, Pearl Dulawan, Amy Hoare, Caroline Jolley, Abigail Knighton, M. Malim, Sheetal Patale, Ida Peralta, Natassia Powell, Albert Ramos, K. Shevket, Fabio Speranza & Amelie Te
Guy’s and St Thomas’ NHS Foundation Trust, London, UK
Laura Aguilar Jimenez, Gill Arbane, Sarah Betts, Karen Bisnauthsing, A. Dewar, Nicholas Hart, G. Kaltsakas, Helen Kerslake, Murphy Magtoto, Philip Marino, L. M. Martinez, Marlies Ostermann, Jennifer Rossdale & Teresa Solano
Royal Free London NHS Foundation Trust, London, UK
Shanaz Ahmad, Simon Brill, John Hurst, Hannah Jarvis, C. Laing, Lai Lim, S. Mandal, Darwin Matila, Olaoluwa Olaosebikan & Claire Singh
University Hospital Birmingham NHS Foundation Trust and University of Birmingham, Birmingham, UK
N. Ahmad Haider, Catherine Atkin, Rhiannon Baggott, Michelle Bates, A. Botkai, Anna Casey, B. Cooper, Joanne Dasgin, Camilla Dawson, Katharine Draxlbauer, N. Gautam, J. Hazeldine, T. Hiwot, Sophie Holden, Karen Isaacs, T. Jackson, Vicky Kamwa, D. Lewis, Janet Lord, S. Madathil, C. McGee, K. Mcgee, Aoife Neal, Alex Newton-Cox, Joseph Nyaboko, Dhruv Parekh, Z. Peterkin, H. Qureshi, Liz Ratcliffe, Elizabeth Sapey, J. Short, Tracy Soulsby, J. Stockley, Zehra Suleiman, Tamika Thompson, Maximina Ventura, Sinead Walder, Carly Welch, Daisy Wilson, S. Yasmin & Kay Por Yip
Stroke Association, London, UK
Rubina Ahmed & Richard Francis
University College London Hospital and University College London, London, UK
Nyarko Ahwireng, Dongchun Bang, Donna Basire, Jeremy Brown, Rachel Chambers, A. Checkley, R. Evans, M. Heightman, T. Hillman, Joseph Jacob, Roman Jastrub, M. Lipman, S. Logan, D. Lomas, Marta Merida Morillas, Hannah Plant, Joanna Porter, K. Roy & E. Wall
Oxford University Hospitals NHS Foundation Trust and University of Oxford, Oxford, UK
Mark Ainsworth, Asma Alamoudi, Angela Bloss, Penny Carter, M. Cassar, Jin Chen, Florence Conneh, T. Dong, Ranuromanana Evans, V. Ferreira, Emily Fraser, John Geddes, F. Gleeson, Paul Harrison, May Havinden-Williams, P. Jezzard, Ivan Koychev, Prathiba Kurupati, H. McShane, Clare Megson, Stefan Neubauer, Debby Nicoll, C. Nikolaidou, G. Ogg, Edmund Pacpaco, M. Pavlides, Yanchun Peng, Nayia Petousi, John Pimm, Najib Rahman, M. J. Rowland, Kathryn Saunders, Michael Sharpe, Nick Talbot, E. M. Tunnicliffe & C. Xie
St George’s University Hospitals NHS Foundation Trust, London, UK
Mariam Ali, Raminder Aul, A. Dunleavy, D. Forton, Mark Mencias, N. Msimanga, T. Samakomva, Sulman Siddique, Vera Tavoukjian & J. Teixeira
University Hospitals of Leicester NHS Trust and University of Leicester, Leicester, UK
M. Aljaroof, Natalie Armstrong, H. Arnold, Hnin Aung, Majda Bakali, M. Bakau, E. Baldry, Molly Baldwin, Charlotte Bourne, Michelle Bourne, Nigel Brunskill, P. Cairns, Liesel Carr, Amanda Charalambou, C. Christie, Melanie Davies, Enya Daynes, Sarah Diver, Rachael Dowling, Sarah Edwards, C. Edwardson, H. Evans, J. Finch, Sarah Glover, Nicola Goodman, Bibek Gooptu, Kate Hadley, Pranab Haldar, Beverley Hargadon, W. Ibrahim, L. Ingram, Kamlesh Khunti, A. Lea, D. Lee, Gerry McCann, P. McCourt, Teresa Mcnally, George Mills, Will Monteiro, Manish Pareek, S. Parker, Anne Prickett, I. N. Qureshi, A. Rowland, Richard Russell, Salman Siddiqui, Sally Singh, J. Skeemer, M. Soares, E. Stringer, T. Thornton, Martin Tobin, T. J. C. Ward, F. Woodhead, Tom Yates & A. J. Yousuf
University of Exeter, Exeter, UK
Louise Allan, Clive Ballard & Andrew McGovern
University of Leicester, Leicester, UK
Richard Allen, Michelle Bingham, Terry Brugha, Selina Finney, Rob Free, Don Jones, Claire Lawson, Daniel Lozano-Rojas, Gardiner Lucy, Alistair Moss, Elizabeta Mukaetova-Ladinska, Petr Novotny, Kimon Ntotsis, Charlotte Overton, John Pearl, Tatiana Plekhanova, M. Richardson, Nilesh Samani, Jack Sargant, Ruth Saunders, M. Sharma, Mike Steiner, Chris Taylor, Sarah Terry, C. Tong, E. Turner, J. Wormleighton & Bang Zhao
Liverpool University Hospitals NHS Foundation Trust and University of Liverpool, Liverpool, UK
Lisa Allerton, Ann Marie Allt, M. Beadsworth, Anthony Berridge, Jo Brown, Shirley Cooper, Andy Cross, Sylviane Defres, S. L. Dobson, Joanne Earley, N. French, Kera Hainey, Hayley Hardwick, Jenny Hawkes, Victoria Highett, Sabina Kaprowska, Angela Key, Lara Lavelle-Langham, N. Lewis-Burke, Gladys Madzamba, Flora Malein, Sophie Marsh, Chloe Mears, Lucy Melling, Matthew Noonan, L. Poll, James Pratt, Emma Richardson, Anna Rowe, Victoria Shaw, K. A. Tripp, Lilian Wajero, S. A. Williams-Howard, Dan Wootton & J. Wyles
Sherwood Forest Hospitals NHS Foundation Trust, Nottingham, UK
Lynne Allsop, Kaytie Bennett, Phil Buckley, Margaret Flynn, Mandy Gill, Camelia Goodwin, M. Greatorex, Heidi Gregory, Cheryl Heeley, Leah Holloway, Megan Holmes, John Hutchinson, Jill Kirk, Wayne Lovegrove, Terri Ann Sewell, Sarah Shelton, D. Sissons, Katie Slack, Susan Smith, D. Sowter, Sarah Turner, V. Whitworth & Inez Wynter
Nottingham University Hospitals NHS Trust and University of Nottingham, London, UK
Paula Almeida, Akram Hosseini, Robert Needham & Karen Shaw
Manchester University NHS Foundation Trust and University of Manchester, London, UK
Bashar Al-Sheklly, Cristina Avram, John Blaikely, M. Buch, N. Choudhury, David Faluyi, T. Felton, T. Gorsuch, Neil Hanley, Tracy Hussell, Zunaira Kausar, Natasha Odell, Rebecca Osbourne, Karen Piper Hanley, K. Radhakrishnan & Sue Stockdale
Imperial College London, London, UK
Danny Altmann, Anew Frankel, Luke S. Howard, Desmond Johnston, Liz Lightstone, Anne Lingford-Hughes, William Man, Steve McAdoo, Jane Mitchell, Philip Molyneaux, Christos Nicolaou, D. P. O’Regan, L. Price, Jennifer K. Quint, David Smith, Jonathon Valabhji, Simon Walsh, Martin Wilkins & Michelle Willicombe
Hampshire Hospitals NHS Foundation Trust, Basingstoke, UK
Maria Alvarez Corral, Ava Maria Arias, Emily Bevan, Denise Griffin, Jane Martin, J. Owen, Sheila Payne, A. Prabhu, Annabel Reed, Will Storrar, Nick Williams & Caroline Wrey Brown
British Heart Foundation, Birmingham, UK
Shannon Amoils
NHS Greater Glasgow and Clyde Health Board and University of Glasgow, Glasgow, UK
David Anderson, Neil Basu, Hannah Bayes, Colin Berry, Ammani Brown, Andrew Dougherty, K. Fallon, L. Gilmour, D. Grieve, K. Mangion, I. B. McInnes, A. Morrow, Kathryn Scott & R. Sykes
University of Oxford, Oxford, UK
Charalambos Antoniades, A. Bates, M. Beggs, Kamaldeep Bhui, Katie Breeze, K. M. Channon, David Clark, X. Fu, Masud Husain, Lucy Kingham, Paul Klenerman, Hanan Lamlum, X. Li, E. Lukaschuk, Celeste McCracken, K. McGlynn, R. Menke, K. Motohashi, T. E. Nichols, Godwin Ogbole, S. Piechnik, I. Propescu, J. Propescu, A. A. Samat, Z. B. Sanders, Louise Sigfrid & M. Webster
Belfast Health and Social Care Trust and Queen’s University Belfast, Belfast, UK
Cherie Armour, Vanessa Brown, John Busby, Bronwen Connolly, Thelma Craig, Stephen Drain, Liam Heaney, Bernie King, Nick Magee, E. Major, Danny McAulay, Lorcan McGarvey, Jade McGinness, Tunde Peto & Roisin Stone
Airedale NHS Foundation Trust, Keighley, UK
Lisa Armstrong, Brigid Hairsine, Helen Henson, Claire Kurasz, Alison Shaw & Liz Shenton
Wrightington Wigan and Leigh NHS Trust, Wigan, UK
A. Ashish, Josh Cooper & Emma Robinson
Leeds Teaching Hospitals and University of Leeds, Leeds, UK
Andrew Ashworth, Paul Beirne, Jude Clarke, C. Coupland, Matthhew Dalton, Clair Favager, Jodie Glossop, John Greenwood, Lucy Hall, Tim Hardy, Amy Humphries, Jennifer Murira, Dan Peckham, S. Plein, Jade Rangeley, Gwen Saalmink, Ai Lyn Tan, Elaine Wade, Beverley Whittam, Nicola Window & Janet Woods
University of Liverpool, Liverpool, UK
M. Ashworth, D. Cuthbertson, G. Kemp, Anne McArdle, Benedict Michael, Will Reynolds, Lisa Spencer, Ben Vinson, Katie A. Ahmed, Jane A. Armstrong, Milton Ashworth, Innocent G. Asiimwe, Siddharth Bakshi, Samantha L. Barlow, Laura Booth, Benjamin Brennan, Katie Bullock, Nicola Carlucci, Emily Cass, Benjamin W. A. Catterall, Jordan J. Clark, Emily A. Clarke, Sarah Cole, Louise Cooper, Helen Cox, Christopher Davis, Oslem Dincarslan, Alejandra Doce Carracedo, Chris Dunn, Philip Dyer, Angela Elliott, Anthony Evans, Lorna Finch, Lewis W. S. Fisher, Lisa Flaherty, Terry Foster, Isabel Garcia-Dorival, Philip Gunning, Catherine Hartley, Karl Holden, Anthony Holmes, Rebecca L. Jensen, Christopher B. Jones, Trevor R. Jones, Shadia Khandaker, Katharine King, Robyn T. Kiy, Chrysa Koukorava, Annette Lake, Suzannah Lant, Diane Latawiec, Lara Lavelle-Langham, Daniella Lefteri, Lauren Lett, Lucia A. Livoti, Maria Mancini, Hannah Massey, Nicole Maziere, Sarah McDonald, Laurence McEvoy, John McLauchlan, Soeren Metelmann, Nahida S. Miah, Joanna Middleton, Joyce Mitchell, Ellen G. Murphy, Rebekah Penrice-Randal, Jack Pilgrim, Tessa Prince, P. Matthew Ridley, Debby Sales, Rebecca K. Shears, Benjamin Small, Krishanthi S. Subramaniam, Agnieska Szemiel, Aislynn Taggart, Jolanta Tanianis-Hughes, Jordan Thomas, Erwan Trochu, Libby van Tonder, Eve Wilcock & J. Eunice Zhang
University College London, London, UK
Shahab Aslani, Amita Banerjee, R. Batterham, Gabrielle Baxter, Robert Bell, Anthony David, Emma Denneny, Alun Hughes, W. Lilaonitkul, P. Mehta, Ashkan Pakzad, Bojidar Rangelov, B. Williams, James Willoughby & Moucheng Xu
Hull University Teaching Hospitals NHS Trust and University of Hull, Hull, UK
Paul Atkin, K. Brindle, Michael Crooks, Katie Drury, Nicholas Easom, Rachel Flockton, L. Holdsworth, A. Richards, D. L. Sykes, Susannah Thackray-Nocera & C. Wright
East Kent Hospitals University NHS Foundation Trust, Canterbury, UK
Liam Austin, Eva Beranova, Tracey Cosier, Joanne Deery, Tracy Hazelton, Carly Price, Hazel Ramos, Reanne Solly, Sharon Turney & Heather Weston
Baillie Gifford Pandemic Science Hub, Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
Nikos Avramidis, J. Kenneth Baillie, Erola Pairo-Castineira & Konrad Rawlik
Roslin Institute, University of Edinburgh, Edinburgh, UK
Nikos Avramidis, J. Kenneth Baillie & Erola Pairo-Castineira
Newcastle upon Tyne Hospitals NHS Foundation Trust and University of Newcastle, Newcastle upon Tyne, UK
A. Ayoub, J. Brown, G. Burns, Gareth Davies, Anthony De Soyza, Carlos Echevarria, Helen Fisher, C. Francis, Alan Greenhalgh, Philip Hogarth, Joan Hughes, Kasim Jiwa, G. Jones, G. MacGowan, D. Price, Avan Sayer, John Simpson, H. Tedd, S. Thomas, Sophie West, M. Witham, S. Wright & A. Young
East Cheshire NHS Trust, Macclesfield, UK
Marta Babores, Maureen Holland, Natalie Keenan, Sharlene Shashaa & Helen Wassall
Sheffield Teaching NHS Foundation Trust and University of Sheffield, Sheffield, UK
J. Bagshaw, M. Begum, K. Birchall, Robyn Butcher, H. Carborn, Flora Chan, Kerry Chapman, Yutung Cheng, Luke Chetham, Cameron Clark, Zach Coburn, Joby Cole, Myles Dixon, Alexandra Fairman, J. Finnigan, H. Foot, David Foote, Amber Ford, Rebecca Gregory, Kate Harrington, L. Haslam, L. Hesselden, J. Hockridge, Ailsa Holbourn, B. Holroyd-Hind, L. Holt, Alice Howell, E. Hurditch, F. Ilyas, Claire Jarman, Allan Lawrie, Ju Hee Lee, Elvina Lee, Rebecca Lenagh, Alison Lye, Irene Macharia, M. Marshall, Angeline Mbuyisa, J. McNeill, Sharon Megson, J. Meiring, L. Milner, S. Misra, Helen Newell, Tom Newman, C. Norman, Lorenza Nwafor, Dibya Pattenadk, Megan Plowright, Julie Porter, Phillip Ravencroft, C. Roddis, J. Rodger, Peter Saunders, J. Sidebottom, Jacqui Smith, Laurie Smith, N. Steele, G. Stephens, R. Stimpson, B. Thamu, N. Tinker, Kim Turner, Helena Turton, Phillip Wade, S. Walker, James Watson, Imogen Wilson & Amira Zawia
University of Nottingham, Nottingham, UK
David Baguley, Chris Coleman, E. Cox, Laura Fabbri, Susan Francis, Ian Hall, E. Hufton, Simon Johnson, Fasih Khan, Paaig Kitterick, Richard Morriss, Nick Selby, Iain Stewart & Louise Wright
Wirral University Teaching Hospital, Wirral, UK
Elisabeth Bailey, Anne Reddington & Andrew Wight
MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Edinburgh, UK
University of Swansea, Swansea, UK
University of Southampton, London, UK
David Baldwin, P. C. Calder, Nathan Huneke & Gemma Simons
Royal Brompton and Harefield Clinical Group, Guy’s and St Thomas’ NHS Foundation Trust, London, UK
R. E. Barker, Daniele Cristiano, N. Dormand, P. George, Mahitha Gummadi, S. Kon, Kamal Liyanage, C. M. Nolan, B. Patel, Suhani Patel, Oliver Polgar, L. Price, P. Shah, Suver Singh & J. A. Walsh
York and Scarborough NHS Foundation Trust, York, UK
Laura Barman, Claire Brookes, K. Elliott, L. Griffiths, Zoe Guy, Kate Howard, Diana Ionita, Heidi Redfearn, Carol Sarginson & Alison Turnbull
NHS Highland, Inverness, UK
Fiona Barrett, A. Donaldson & Beth Sage
Royal Papworth Hospital NHS Foundation Trust, Cambridge, UK
Helen Baxendale, Lucie Garner, C. Johnson, J. Mackie, Alice Michael, J. Newman, Jamie Pack, K. Paques, H. Parfrey, J. Parmar & A. Reddy
University Hospitals of Derby and Burton, Derby, UK
Paul Beckett, Caroline Dickens & Uttam Nanda
NHS Lanarkshire, Hamilton, UK
Murdina Bell, Angela Brown, M. Brown, R. Hamil, Karen Leitch, L. Macliver, Manish Patel, Jackie Quigley, Andrew Smith & B. Welsh
Cambridge University Hospitals NHS Foundation Trust, NIHR Cambridge Clinical Research Facility and University of Cambridge, Cambridge, UK
Areti Bermperi, Isabel Cruz, K. Dempsey, Anne Elmer, Jonathon Fuld, H. Jones, Sherly Jose, Stefan Marciniak, M. Parkes, Carla Ribeiro, Jessica Taylor, Mark Toshner, L. Watson & J. Worsley
Loughborough University, Loughborough, UK
Lettie Bishop & David Stensel
Betsi Cadwallader University Health Board, Bangor, UK
Annette Bolger, Ffyon Davies, Ahmed Haggar, Joanne Lewis, Arwel Lloyd, R. Manley, Emma McIvor, Daniel Menzies, K. Roberts, W. Saxon, David Southern, Christian Subbe & Victoria Whitehead
Nottingham University Hospitals NHS Trust and University of Nottingham, Nottingham, UK
Charlotte Bolton, J. Bonnington, Melanie Chrystal, Catherine Dupont, Paul Greenhaff, Ayushman Gupta, W. Jang, S. Linford, Laura Matthews, Athanasios Nikolaidis, Sabrina Prosper & Andrew Thomas
King’s College London, London, UK
Kate Bramham, M. Brown, Khalida Ismail, Tim Nicholson, Carmen Pariante, Claire Sharpe, Simon Wessely & J. Whitney
Bradford Teaching Hospitals NHS Foundation Trust, Bradford, UK
Lucy Brear, Karen Regan, Dinesh Saralaya & Kim Storton
South London and Maudsley NHS Foundation Trust and King’s College London, London, UK
G. Breen & M. Hotopf
London School of Hygiene and Tropical Medicine, London, UK
Andrew Briggs
Whittington Health NHS Trust, London, UK
E. Bright, P. Crisp, Ruvini Dharmagunawardena & M. Stern
Cardiff and Vale University Health Board, Cardiff, UK
Lauren Broad, Teriann Evans, Matthew Haynes, L. Jones, Lucy Knibbs, Alison McQueen, Catherine Oliver, Kerry Paradowski, Ramsey Sabit & Jenny Williams
Yeovil District Hospital NHS Foundation Trust, Yeovil, UK
Andrew Broadley
University of Birmingham, Birmingham, UK
Mattew Broome, Paul McArdle, Paul Moss, David Thickett, Rachel Upthegrove, Dan Wilkinson, David Wraith & Erin L. Aldera
BHF Centre for Cardiovascular Science, University of Edinburgh, Edinburgh, UK
Anda Bularga
University of Cambridge, Cambridge, UK
Ed Bullmore, Jonathon Heeney, Claudia Langenberg, William Schwaeble, Charlotte Summers & J. Weir McCall
NIHR Leicester Biomedical Research Centre–Respiratory Patient and Public Involvement Group, Leicester, UK
Jenny Bunker, Rhyan Gill & Rashmita Nathu
Imperial College Healthcare NHS Trust and Imperial College London, London, UK
L. Burden, Ellen Calvelo, Bethany Card, Caitlin Carr, Edwin Chilvers, Donna Copeland, P. Cullinan, Patrick Daly, Lynsey Evison, Tamanah Fayzan, Hussain Gordon, Sulaimaan Haq, Gisli Jenkins, Clara King, Onn Min Kon, Katherine March, Myril Mariveles, Laura McLeavey, Silvia Moriera, Unber Munawar, Uchechi Nwanguma, Lorna Orriss-Dib, Alexandra Ross, Maura Roy, Emily Russell, Katherine Samuel, J. Schronce, Neil Simpson, Lawrence Tarusan, David Thomas, Chloe Wood & Najira Yasmin
Harrogate and District NHD Foundation Trust, Harrogate, UK
Tracy Burdett, James Featherstone, Cathy Lawson, Alison Layton, Clare Mills & Lorraine Stephenson
Newcastle University/Chair of NIHR Dementia TRC, Newcastle, UK
Oxford University Hospitals NHS Foundation Trust, Oxford, UK
A. Burns & N. Kanellakis
Tameside and Glossop Integrated Care NHS Foundation Trust, Ashton-under-Lyne, UK
Al-Tahoor Butt, Martina Coulding, Heather Jones, Susan Kilroy, Jacqueline McCormick, Jerome McIntosh, Heather Savill, Victoria Turner & Joanne Vere
University of Oxford, Nuffield Department of Medicine, Oxford, UK
University of Glasgow, Glasgow, UK
Jonathon Cavanagh, S. MacDonald, Kate O’Donnell, John Petrie, Naveed Sattar & Mark Spears
United Lincolnshire Hospitals NHS Trust, Grantham, UK
Manish Chablani & Lynn Osborne
Department of Psychological Medicine, Institute of Psychiatry, Psychology and Neuroscience, King’s College London, London, UK
Trudie Chalder
University Hospital of South Manchester NHS Foundation Trust, Manchester, UK
N. Chaudhuri
University Hospital Southampton NHS Foundation Trust and University of Southampton, Southampton, UK
Caroline Childs, R. Djukanovic, S. Fletcher, Matt Harvey, Mark Jones, Elizabeth Marouzet, B. Marshall, Reena Samuel, T. Sass, Tim Wallis & Helen Wheeler
King’s College Hospital/Guy’s and St Thomas’ NHS FT, London, UK
A. Chiribiri & C. O’Brien
Barts Health NHS Trust, London, UK
K. Chong-James, C. David, W. Y. James, Paul Pfeffer & O. Zongo
NHS Lothian and University of Edinburgh, Edinburgh, UK
Gaunab Choudhury, S. Clohisey, Andrew Deans, J. Furniss, Ewen Harrison, S. Kelly & Aziz Sheikh
School of Cardiovascular Medicine and Sciences. King’s College London, London, UK
Phillip Chowienczyk
Lewisham and Greenwich NHS Trust, London, UK
Hywel Dda University Health Board, Haverfordwest, UK
S. Coetzee, Kim Davies, Rachel Ann Hughes, Ronda Loosley, Heather McGuinness, Abdelrahman Mohamed, Linda O’Brien, Zohra Omar, Emma Perkins, Janet Phipps, Gavin Ross, Abigail Taylor, Helen Tench & Rebecca Wolf-Roberts
NHS Tayside and University of Dundee, Dundee, UK
David Connell, C. Deas, Anne Elliott, J. George, S. Mohammed, J. Rowland, A. R. Solstice, Debbie Sutherland & Caroline Tee
Swansea Bay University Health Board, Port Talbot, UK
Lynda Connor, Amanda Cook, Gwyneth Davies, Tabitha Rees, Favas Thaivalappil & Caradog Thomas
Faculty of Medicine, Nursing and Health Sciences, School of Biomedical Sciences, Monash University, Melbourne, Victoria, Australia
Eamon Coughlan
Rotherham NHS Foundation Trust, Rotherham, UK
Alison Daniels, Anil Hormis, Julie Ingham & Lisa Zeidan
Salford Royal NHS Foundation Trust, Salford, UK
P. Dark, Nawar Diar-Bakerly, D. Evans, E. Hardy, Alice Harvey, D. Holgate, Sean Knight, N. Mairs, N. Majeed, L. McMorrow, J. Oxton, Jessica Pendlebury, C. Summersgill, R. Ugwuoke & S. Whittaker
Cwm Taf Morgannwg University Health Board, Mountain Ash, UK
Ellie Davies, Cerys Evenden, Alyson Hancock, Kia Hancock, Ceri Lynch, Meryl Rees, Lisa Roche, Natalie Stroud & T. Thomas-Woods
Borders General Hospital, NHS Borders, Melrose, UK
Joy Dawson, Hosni El-Taweel & Leanne Robinson
Aneurin Bevan University Health Board, Caerleon, UK
Amanda Dell, Sara Fairbairn, Nancy Hawkings, Jill Haworth, Michaela Hoare, Victoria Lewis, Alice Lucey, Georgia Mallison, Heeah Nassa, Chris Pennington, Andrea Price, Claire Price, Andrew Storrie, Gemma Willis & Susan Young
University of Exeter Medical School, Exeter, UK
London North West University Healthcare NHS Trust, London, UK
Shalin Diwanji, Sambasivarao Gurram, Padmasayee Papineni, Sheena Quaid, Gerlynn Tiongson & Ekaterina Watson
Alzheimer’s Research UK, Cambridge, UK
Hannah Dobson
Health and Care Research Wales, Cardiff, UK
Yvette Ellis
University of Bristol, Bristol, UK
Jonathon Evans
University of Sheffield, Sheffield, UK
L. Finnigan, Laura Saunders & James Wild
Great Western Hospital Foundation Trust, Swindon, UK
Eva Fraile & Jacinta Ugoji
Royal Devon and Exeter NHS Trust, Barnstaple, UK
Michael Gibbons
Kettering General Hospital NHS Trust, Kettering, UK
Anne-Marie Guerdette, Melanie Hewitt, R. Reddy, Katie Warwick & Sonia White
NIHR Leicester Biomedical Research Centre, Leicester, UK
Beatriz Guillen-Guio
University of Leeds, Leeds, UK
Elspeth Guthrie & Max Henderson
Royal Surrey NHS Foundation Trust, Cranleigh, UK
Mark Halling-Brown & Katherine McCullough
Chesterfield Royal Hospital NHS Trust, Calow, UK
Edward Harris & Claire Sampson
Long Covid Support, London, UK
Claire Hastie, Natalie Rogers & Nikki Smith
King’s College Hospital, NHS Foundation Trust and King’s College London, London, UK
Department of Oncology and Metabolism, University of Sheffield, Sheffield, UK
Simon Heller
NIHR Office for Clinical Research Infrastructure, London, UK
Katie Holmes
Asthma UK and British Lung Foundation Partnership, London, UK
Ian Jarrold & Samantha Walker
North Middlesex University Hospital NHS Trust, London, UK
Bhagy Jayaraman & Tessa Light
Action for Pulmonary Fibrosis, Peterborough, UK
Cardiff University, National Centre for Mental Health, Cardiff, UK
McPin Foundation, London, UK
Thomas Kabir
Roslin Institute, The University of Edinburgh, Edinburgh, UK
Steven Kerr
The Hillingdon Hospitals NHS Foundation Trust, London, UK
Samantha Kon, G. Landers, Harpreet Lota, Mariam Nasseri & Sofiya Portukhay
Queen Mary University of London, London, UK
Ania Korszun
Swansea University, Swansea Welsh Network, Hywel Dda University Health Board, Swansea, UK
Royal Infirmary of Edinburgh, NHS Lothian, Edinburgh, UK
Nazir I. Lone
Barts Heart Centre, London, UK
Barts Health NHS Trust and Queen Mary University of London, London, UK
Adrian Martineau
Salisbury NHS Foundation Trust, Salisbury, UK
Wadzanai Matimba-Mupaya & Sophia Strong-Sheldrake
University of Newcastle, Newcastle, UK
Hamish McAllister-Williams, Stella-Maria Paddick, Anthony Rostron & John Paul Taylor
Gateshead NHS Trust, Gateshead, UK
W. McCormick, Lorraine Pearce, S. Pugmire, Wendy Stoker & Ann Wilson
Manchester Centre for Clinical Neurosciences, Salford Royal NHS Foundation Trust, Manchester, UK
Katherine McIvor
Kidney Research UK, Peterborough, UK
Aisling McMahon
NHS Dumfries and Galloway, Dumfries, UK
Michael McMahon & Paula Neill
Swansea University, Swansea, UK
MQ Mental Health Research, London, UK
Lea Milligan
BHF Centre for Cardiovascular Science, Usher Institute of Population Health Sciences and Informatics, University of Edinburgh, Edinburgh, UK
Nicholas Mills
Shropshire Community Health NHS Trust, Shropshire, UK
Sharon Painter, Johanne Tomlinson & Louise Warburton
Somerset NHS Foundation Trust, Taunton, UK
Sue Palmer, Dawn Redwood, Jo Tilley, Carinna Vickers & Tania Wainwright
Francis Crick Institute, London, UK
Markus Ralser
Manchester University NHD Foundation Trust, Manchester, UK
Pilar Rivera-Ortega
Diabetes UK, University of Glasgow, Glasgow, UK
Elizabeth Robertson
Barnsley Hospital NHS Foundation Trust, Barnsley, UK
Amy Sanderson
MRC–University of Glasgow Centre for Virus Research, Glasgow, UK
Janet Scott
Diabetes UK, London, UK
Kamini Shah
British Heart Foundation Centre, King’s College London, London, UK
King’s College Hospital NHS Foundation Trust, London, UK
University Hospitals Birmingham NHS Foundation Trust and University of Birmingham, Birmingham, UK
Institute of Cardiovascular and Medical Sciences, BHF Glasgow Cardiovascular Research Centre, University of Glasgow, Glasgow, UK
University College London NHS Foundation Trust, London and Barts Health NHS Trust, London, UK
Northumbria University, Newcastle upon Tyne, UK
Ioannis Vogiatzis
Swansea University and Swansea Welsh Network, Swansea, UK
N. Williams
DUK | NHS Digital, Salford Royal Foundation Trust, Salford, UK
Queen Alexandra Hospital, Portsmouth, UK
- Kayode Adeniji
Princess Royal Hospital, Haywards Heath, UK
Daniel Agranoff & Chi Eziefula
Bassetlaw Hospital, Bassetlaw, UK
Darent Valley Hospital, Dartford, UK
Queen Elizabeth the Queen Mother Hospital, Margate, UK
Ana Alegria
School of Informatics, University of Edinburgh, Edinburgh, UK
Beatrice Alex, Benjamin Bach & James Scott-Brown
North East and North Cumbria Ingerated, Newcastle upon Tyne, UK
Section of Biomolecular Medicine, Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Petros Andrikopoulos, Kanta Chechi, Marc-Emmanuel Dumas, Julian Griffin, Sonia Liggi & Zoltan Takats
Section of Genomic and Environmental Medicine, Respiratory Division, National Heart and Lung Institute, Imperial College London, London, UK
Petros Andrikopoulos, Marc-Emmanuel Dumas, Michael Olanipekun & Anthonia Osagie
John Radcliffe Hospital, Oxford, UK
Brian Angus
Royal Albert Edward Infirmary, Wigan, UK
Abdul Ashish
Manchester Royal Infirmary, Manchester, UK
Dougal Atkinson
MRC Human Genetics Unit, Institute of Genetics and Cancer, University of Edinburgh, Western General Hospital, Crewe Road, Edinburgh, UK
Section of Molecular Virology, Imperial College London, London, UK
Wendy S. Barclay
Furness General Hospital, Barrow-in-Furness, UK
Shahedal Bari
Hull University Teaching Hospital Trust, Kingston upon Hull, UK
Gavin Barlow
Hillingdon Hospital, Hillingdon, UK
Stella Barnass
St Thomas’ Hospital, London, UK
Nicholas Barrett
Coventry and Warwickshire, Coventry, UK
Christopher Bassford
St Michael’s Hospital, Bristol, UK
Sneha Basude
Stepping Hill Hospital, Stockport, UK
David Baxter
Royal Liverpool University Hospital, Liverpool, UK
Michael Beadsworth
Bristol Royal Hospital Children’s, Bristol, UK
Jolanta Bernatoniene
Scarborough Hospital, Scarborough, UK
John Berridge
Golden Jubilee National Hospital, Clydebank, UK
Colin Berry
Liverpool Heart and Chest Hospital, Liverpool, UK
Nicola Best
Centre for Inflammation Research, The Queen’s Medical Research Institute, University of Edinburgh, Edinburgh, UK
Debby Bogaert & Clark D. Russell
James Paget University Hospital, Great Yarmouth, UK
Pieter Bothma & Darell Tupper-Carey
Aberdeen Royal Infirmary, Aberdeen, UK
Robin Brittain-Long
Adamson Hospital, Cupar, UK
Naomi Bulteel
Royal Devon and Exeter Hospital, Exeter, UK
Worcestershire Royal Hospital, Worcester, UK
Andrew Burtenshaw
ISARIC Global Support Centre, Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Gail Carson, Laura Merson & Louise Sigfrid
Conquest Hospital, Hastings, UK
Vikki Caruth
The James Cook University Hospital, Middlesbrough, UK
David Chadwick
Dorset County Hospital, Dorchester, UK
Duncan Chambler
Antimicrobial Resistance and Hospital Acquired Infection Department, Public Health England, London, UK
Meera Chand
Department of Epidemiology and Biostatistics, School of Public Health, Faculty of Medicine, Imperial College London, London, UK
Kanta Chechi
Royal Bournemouth General Hospital, Bournemouth, UK
Harrogate Hospital, Harrogate, UK
Jenny Child
Royal Blackburn Teaching Hospital, Blackburn, UK
Srikanth Chukkambotla
Edinburgh Clinical Research Facility, University of Edinburgh, Edinburgh, UK
Richard Clark, Audrey Coutts, Lorna Donelly, Angie Fawkes, Tammy Gilchrist, Katarzyna Hafezi, Louise MacGillivray, Alan Maclean, Sarah McCafferty, Kirstie Morrice, Lee Murphy & Nicola Wrobel
Torbay Hospital, Torquay, UK
Northern General Hospital, Sheffield, UK
Paul Collini, Cariad Evans & Gary Mills
Liverpool Clinical Trials Centre, University of Liverpool, Liverpool, UK
Marie Connor, Jo Dalton, Chloe Donohue, Carrol Gamble, Michelle Girvan, Sophie Halpin, Janet Harrison, Clare Jackson, Laura Marsh, Stephanie Roberts & Egle Saviciute
Department of Infectious Disease, Imperial College London, London, UK
Graham S. Cooke & Shiranee Sriskandan
St Georges Hospital (Tooting), London, UK
Catherine Cosgrove
Blackpool Victoria Hospital, Blackpool, UK
Jason Cupitt & Joanne Howard
The Royal London Hospital, London, UK
Maria-Teresa Cutino-Moguel
MRC-University of Glasgow Centre for Virus Research, Glasgow, UK
Ana da Silva Filipe, Antonia Y. W. Ho, Sarah E. McDonald, Massimo Palmarini, David L. Robertson, Janet T. Scott & Emma C. Thomson
Salford Royal Hospital, Salford, UK
University Hospital of North Durham, Durham, UK
Chris Dawson
Norfolk and Norwich University Hospital, Norwich, UK
Samir Dervisevic
Intensive Care Unit, Royal Infirmary Edinburgh, Edinburgh, UK
Annemarie B. Docherty & Seán Keating
Institute of Infection, Veterinary and Ecological Sciences, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, UK
Cara Donegan & Rebecca G. Spencer
Salisbury District Hospital, Salisbury, UK
Phil Donnison
National Phenome Centre, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Gonçalo dos Santos Correia, Matthew Lewis, Lynn Maslen, Caroline Sands, Zoltan Takats & Panteleimon Takis
Section of Bioanalytical Chemistry, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Gonçalo dos Santos Correia, Matthew Lewis, Lynn Maslen, Caroline Sands & Panteleimon Takis
Guy’s and St Thomas’, NHS Foundation Trust, London, UK
Sam Douthwaite, Michael MacMahon, Marlies Ostermann & Manu Shankar-Hari
The Royal Oldham Hospital, Oldham, UK
Andrew Drummond
European Genomic Institute for Diabetes, Institut Pasteur de Lille, Lille University Hospital, University of Lille, Lille, France
Marc-Emmanuel Dumas
McGill University and Genome Quebec Innovation Centre, Montreal, Qeubec, Canada
National Infection Service, Public Health England, London, UK
Jake Dunning & Maria Zambon
Hereford Count Hospital, Hereford, UK
Ingrid DuRand
Southampton General Hospital, Southampton, UK
Ahilanadan Dushianthan
Northampton General Hospital, Northampton, UK
Tristan Dyer
University Hospital of Wales, Cardiff, UK
Chrisopher Fegan
University Hospitals Bristol NHS Foundation Trust, Bristol, UK
Liverpool School of Tropical Medicine, Liverpool, UK
Tom Fletcher
Leighton Hospital, Crewe, UK
Duncan Fullerton & Elijah Matovu
Manor Hospital, Walsall, UK
Scunthorpe Hospital, Scunthorpe, UK
Sanjeev Garg
Cambridge University Hospital, Cambridge, UK
Effrossyni Gkrania-Klotsas
West Suffolk NHS Foundation Trust, Bury St Edmunds, UK
Basingstoke and North Hampshire Hospital, Basingstoke, UK
Arthur Goldsmith
North Cumberland Infirmary, Carlisle, UK
Clive Graham
Paediatric Liver, GI and Nutrition Centre and MowatLabs, King’s College Hospital, London, UK
Tassos Grammatikopoulos
Institute of Liver Studies, King’s College London, London, UK
Institute of Microbiology and Infection, University of Birmingham, Birmingham, UK
Christopher A. Green
Department of Molecular and Clinical Cancer Medicine, University of Liverpool, Liverpool, UK
William Greenhalf
Institute for Global Health, University College London, London, UK
Rishi K. Gupta
NIHR Health Protection Research Unit, Institute of Infection, Veterinary and Ecological Sciences, Faculty of Health and Life Sciences, University of Liverpool, Liverpool, UK
Hayley Hardwick, Malcolm G. Semple, Tom Solomon & Lance C. W. Turtle
Warwick Hospital, Warwick, UK
Elaine Hardy
Birmingham Children’s Hospital, Birmingham, UK
Stuart Hartshorn
Nottingham City Hospital, Nottingham, UK
Daniel Harvey
Glangwili Hospital Child Health Section, Carmarthen, UK
Peter Havalda
Alder Hey Children’s Hospital, Liverpool, UK
Daniel B. Hawcutt
Department of Infectious Diseases, Queen Elizabeth University Hospital, Glasgow, UK
Antonia Y. W. Ho
Bronglais General Hospital, Aberystwyth, UK
Maria Hobrok
Worthing Hospital, Worthing, UK
Luke Hodgson
Centre for Tropical Medicine and Global Health, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Peter W. Horby
Rotheram District General Hospital, Rotheram, UK
Anil Hormis
Virology Reference Department, National Infection Service, Public Health England, Colindale Avenue, London, UK
Samreen Ijaz
Royal Free Hospital, London, UK
Michael Jacobs & Padmasayee Papineni
Homerton Hospital, London, UK
Airedale Hospital, Airedale, UK
Paul Jennings
Basildon Hospital, Basildon, UK
Agilan Kaliappan
The Christie NHS Foundation Trust, Manchester, UK
Vidya Kasipandian
University Hospital Lewisham, London, UK
Stephen Kegg
The Whittington Hospital, London, UK
Michael Kelsey
Southmead Hospital, Bristol, UK
Jason Kendall
Sheffield Childrens Hospital, Sheffield, UK
Caroline Kerrison
Royal United Hospital, Bath, UK
Ian Kerslake
Department of Pharmacology, University of Liverpool, Liverpool, UK
Nuffield Department of Medicine, Peter Medawar Building for Pathogen Research, University of Oxford, Oxford, UK
Paul Klenerman
Translational Gastroenterology Unit, Nuffield Department of Medicine, University of Oxford, Oxford, UK
Public Health Scotland, Edinburgh, UK
Susan Knight, Eva Lahnsteiner & Sarah Tait
Western General Hospital, Edinburgh, UK
Oliver Koch
Southend University Hospital NHS Foundation Trust, Southend-on-Sea, UK
Gouri Koduri
Hinchingbrooke Hospital, Huntingdon, UK
George Koshy & Tamas Leiner
Royal Preston Hospital, Fulwood, UK
Shondipon Laha
University Hospital (Coventry), Coventry, UK
Steven Laird
The Walton Centre, Liverpool, UK
Susan Larkin
ISARIC, Global Support Centre, COVID-19 Clinical Research Resources, Epidemic diseases Research Group, Oxford (ERGO), University of Oxford, Oxford, UK
James Lee & Daniel Plotkin
Centre for Health Informatics, Division of Informatics, Imaging and Data Science, School of Health Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester Academic Health Science Centre, Manchester, UK
Gary Leeming
Hull Royal Infirmary, Hull, UK
Patrick Lillie
Nottingham University Hospitals NHS Trust:, Nottingham, UK
Wei Shen Lim
Darlington Memorial Hospital, Darlington, UK
Queen Elizabeth Hospital (Gateshead), Gateshead, UK
Vanessa Linnett
Warrington Hospital, Warrington, UK
Jeff Little
Bristol Royal Hospital for Children, Bristol, UK
Mark Lyttle
St Mary’s Hospital (Isle of Wight), Isle of Wight, UK
Emily MacNaughton
The Tunbridge Wells Hospital, Royal Tunbridge Wells, UK
Ravish Mankregod
Huddersfield Royal, Huddersfield, UK
Countess of Chester Hospital, Liverpool, UK
Ruth McEwen & Lawrence Wilson
Frimley Park Hospital, Frimley, UK
Manjula Meda
Nuffield Department of Medicine, John Radcliffe Hospital, Oxford, UK
Alexander J. Mentzer
Department of Microbiology/Infectious Diseases, Oxford University Hospitals NHS Foundation Trust, John Radcliffe Hospital, Oxford, UK
MRC Human Genetics Unit, MRC Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK
Alison M. Meynert & Murray Wham
St James University Hospital, Leeds, UK
Jane Minton
Arrowe Park Hospital, Birkenhead, UK
Kavya Mohandas
Great Ormond Street Hospital, London, UK
Royal Shrewsbury Hospital, Shrewsbury, UK
Addenbrookes Hospital, Cambridge, UK
Elinoor Moore
Institute of Infection, Veterinary and Ecological Sciences, University of Liverpool, Liverpool, UK
Shona C. Moore, William A. Paxton & Georgios Pollakis
East Surrey Hospital (Redhill), Redhill, UK
Patrick Morgan
Burton Hospital, Burton, UK
Craig Morris & Tim Reynolds
Peterborough City Hospital, Peterborough, UK
Katherine Mortimore
Kent and Canterbury Hospital, Canterbury, UK
Samuel Moses
Weston Area General Trust, Bristol, UK
Mbiye Mpenge
Bedfordshire Hospital, Bedfordshire, UK
Rohinton Mulla
Glasgow Royal Infirmary, Glasgow, UK
Michael Murphy
Macclesfield General Hospital, Macclesfield, UK
Thapas Nagarajan
Derbyshire Healthcare, Derbyshire, UK
Megan Nagel
Chelsea and Westminster Hospital, London, UK
Mark Nelson & Matthew K. O’Shea
Watford General Hospital, Watford, UK
Lillian Norris & Tom Stambach
EPCC, University of Edinburgh, Edinburgh, UK
Lucy Norris
Section of Biomolecular Medicine, Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, London, UK
Michael Olanipekun
Imperial College Healthcare NHS Trust: London, London, UK
Peter J. M. Openshaw
Division of Systems Medicine, Department of Metabolism, Digestion and Reproduction, Imperial College London, London, UK
Anthonia Osagie
Prince Philip Hospital, Llanelli, UK
Igor Otahal & Andrew Workman
George Eliot Hospital – Acute Services, Nuneaton, UK
Molecular and Clinical Cancer Medicine, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, Liverpool, UK
Carlo Palmieri
Clatterbridge Cancer Centre NHS Foundation Trust, Liverpool, UK
Kettering General Hospital, Kettering, UK
Selva Panchatsharam
University Hospitals of North Midlands NHS Trust, North Midlands, UK
Danai Papakonstantinou
Russells Hall Hospital, Dudley, UK
Hassan Paraiso
Harefield Hospital, Harefield, UK
Lister Hospital, Lister, UK
Natalie Pattison
Musgrove Park Hospital, Taunton, UK
Justin Pepperell
Kingston Hospital, Kingston, UK
Mark Peters
Queen’s Hospital, Romford, UK
Mandeep Phull
Southport and Formby District General Hospital, Southport, UK
Stefania Pintus
St George’s University of London, London, UK
Tim Planche
King’s College Hospital (Denmark Hill), London, UK
Centre for Clinical Infection and Diagnostics Research, Department of Infectious Diseases, School of Immunology and Microbial Sciences, King’s College London, London, UK
Nicholas Price
Department of Infectious Diseases, Guy’s and St Thomas’ NHS Foundation Trust, London, UK
The Clatterbridge Cancer Centre NHS Foundation, Bebington, UK
David Price
The Great Western Hospital, Swindon, UK
Rachel Prout
Ninewells Hospital, Dundee, UK
Nikolas Rae
Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK
Andrew Rambaut
Poole Hospital NHS Trust, Poole, UK
Henrik Reschreiter
William Harvey Hospital, Ashford, UK
Neil Richardson
King’s Mill Hospital, Sutton-in-Ashfield, UK
Mark Roberts
Liverpool Women’s Hospital, Liverpool, UK
Devender Roberts
Pinderfields Hospital, Wakefield, UK
Alistair Rose
North Devon District Hospital, Barnstaple, UK
Guy Rousseau
Queen Elizabeth Hospital, Birmingham, UK
Tameside General Hospital, Ashton-under-Lyne, UK
Brendan Ryan
City Hospital (Birmingham), Birmingham, UK
Taranprit Saluja
Department of Pediatrics and Virology, St Mary’s Medical School Bldg, Imperial College London, London, UK
Vanessa Sancho-Shimizu
The Newcastle Upon Tyne Hospitals NHS Foundation Trust, Newcastle Upon Tyne, UK
Matthias Schmid
NHS Greater Glasgow and Clyde, Glasgow, UK
Janet T. Scott
Respiratory Medicine, Institute in The Park, University of Liverpool, Alder Hey Children’s Hospital, Liverpool, UK
Malcolm G. Semple
Broomfield Hospital, Broomfield, UK
Stoke Mandeville, UK
Prad Shanmuga
University Hospital of North Tees, Stockton-on-Tees, UK
Anil Sharma
Institute of Translational Medicine, University of, Liverpool, Merseyside, UK
Victoria E. Shaw
Royal Manchester Children’s Hospital, Manchester, UK
Anna Shawcross
New Cross Hospital, Wolverhampton, UK
Jagtur Singh Pooni
Bedford Hospital, Bedford, UK
Jeremy Sizer
Colchester General Hospital, Colchester, UK
Richard Smith
University Hospital Birmingham NHS Foundation Trust, Birmingham, UK
Catherine Snelson & Tony Whitehouse
Walton Centre NHS Foundation Trust, Liverpool, UK
Tom Solomon
Chesterfield Royal Hospital, Calow, UK
Nick Spittle
MRC Centre for Molecular Bacteriology and Infection, Imperial College London, London, UK
Shiranee Sriskandan
Princess Alexandra Hospital, Harlow, UK
Nikki Staines & Shico Visuvanathan
Milton Keynes Hospital, Eaglestone, UK
Richard Stewart
Division of Structural Biology, The Wellcome Centre for Human Genetics, University of Oxford, Oxford, UK
David Stuart
Royal Bolton Hopital, Farnworth, UK
Pradeep Subudhi
Department of Medicine, University of Cambridge, Cambridge, UK
Charlotte Summers
Department of Child Life and Health, University of Edinburgh, Edinburgh, UK
Olivia V. Swann
Royal Gwent (Newport), Newport, UK
Tamas Szakmany
The Royal Marsden Hospital (London), London, UK
Kate Tatham
Blood Borne Virus Unit, Virus Reference Department, National Infection Service, Public Health England, London, UK
Richard S. Tedder
Transfusion Microbiology, National Health Service Blood and Transplant, London, UK
Department of Medicine, Imperial College London, London, UK
Queen Victoria Hospital (East Grinstead), East Grinstead, UK
Leeds Teaching Hospitals NHS Trust, Leeds, UK
Robert Thompson
Royal Stoke University Hospital, Stoke-on-Trent, UK
Chris Thompson
Whiston Hospital, Rainhill, UK
Ascanio Tridente
Tropical and Infectious Disease Unit, Royal Liverpool University Hospital, Liverpool, UK
Lance C. W. Turtle
Croydon University Hospital, Thornton Heath, UK
Mary Twagira
Gloucester Royal, Gloucester, UK
Nick Vallotton
West Hertfordshire Teaching Hospitals NHS Trust, Hertfordshire, UK
Rama Vancheeswaran
North Middlesex Hospital, London, UK
Rachel Vincent
Medway Maritime Hospital, Gillingham, UK
Lisa Vincent-Smith
Royal Papworth Hospital Everard, Cambridge, UK
Alan Vuylsteke
Derriford (Plymouth), Plymouth, UK
St Helier Hospital, Sutton, UK
Rachel Wake
Royal Berkshire Hospital, Reading, UK
Andrew Walden
Royal Liverpool Hospital, Liverpool, UK
Ingeborg Welters
Bradford Royal infirmary, Bradford, UK
Paul Whittaker
Central Middlesex, London, UK
Ashley Whittington
Royal Cornwall Hospital (Tresliske), Truro, UK
Meme Wijesinghe
North Bristol NHS Trust, Bristol, UK
Martin Williams
St. Peter’s Hospital, Runnymede, UK
Stephen Winchester
Leicester Royal Infirmary, Leicester, UK
Martin Wiselka
Grantham and District Hospital, Grantham, UK
Adam Wolverson
Aintree University Hospital, Liverpool, UK
Daniel G. Wootton
North Tyneside General Hospital, North Shields, UK
Bryan Yates
Queen Elizabeth Hospital, King’s Lynn, UK
Peter Young
You can also search for this author in PubMed Google Scholar
PHOSP-COVID collaborative group
- Kathryn Abel
- , H. Adamali
- , Davies Adeloye
- , Oluwaseun Adeyemi
- , Rita Adrego
- , Laura Aguilar Jimenez
- , Shanaz Ahmad
- , N. Ahmad Haider
- , Rubina Ahmed
- , Nyarko Ahwireng
- , Mark Ainsworth
- , Asma Alamoudi
- , Mariam Ali
- , M. Aljaroof
- , Louise Allan
- , Richard Allen
- , Lisa Allerton
- , Lynne Allsop
- , Ann Marie Allt
- , Paula Almeida
- , Bashar Al-Sheklly
- , Danny Altmann
- , Maria Alvarez Corral
- , Shannon Amoils
- , David Anderson
- , Charalambos Antoniades
- , Gill Arbane
- , Ava Maria Arias
- , Cherie Armour
- , Lisa Armstrong
- , Natalie Armstrong
- , David Arnold
- , H. Arnold
- , A. Ashish
- , Andrew Ashworth
- , M. Ashworth
- , Shahab Aslani
- , Hosanna Assefa-Kebede
- , Paul Atkin
- , Catherine Atkin
- , Raminder Aul
- , Hnin Aung
- , Liam Austin
- , Cristina Avram
- , Nikos Avramidis
- , Marta Babores
- , Rhiannon Baggott
- , J. Bagshaw
- , David Baguley
- , Elisabeth Bailey
- , J. Kenneth Baillie
- , Steve Bain
- , Majda Bakali
- , E. Baldry
- , Molly Baldwin
- , David Baldwin
- , Clive Ballard
- , Amita Banerjee
- , Dongchun Bang
- , R. E. Barker
- , Laura Barman
- , Perdita Barran
- , Shaney Barratt
- , Fiona Barrett
- , Donna Basire
- , Neil Basu
- , Michelle Bates
- , R. Batterham
- , Helen Baxendale
- , Gabrielle Baxter
- , Hannah Bayes
- , M. Beadsworth
- , Paul Beckett
- , Paul Beirne
- , Murdina Bell
- , Robert Bell
- , Kaytie Bennett
- , Eva Beranova
- , Areti Bermperi
- , Anthony Berridge
- , Colin Berry
- , Sarah Betts
- , Emily Bevan
- , Kamaldeep Bhui
- , Michelle Bingham
- , K. Birchall
- , Lettie Bishop
- , Karen Bisnauthsing
- , John Blaikely
- , Angela Bloss
- , Annette Bolger
- , Charlotte Bolton
- , J. Bonnington
- , A. Botkai
- , Charlotte Bourne
- , Michelle Bourne
- , Kate Bramham
- , Lucy Brear
- , Jonathon Breeze
- , Katie Breeze
- , Andrew Briggs
- , E. Bright
- , Christopher Brightling
- , Simon Brill
- , K. Brindle
- , Lauren Broad
- , Andrew Broadley
- , Claire Brookes
- , Mattew Broome
- , Vanessa Brown
- , Ammani Brown
- , Angela Brown
- , Jeremy Brown
- , Terry Brugha
- , Nigel Brunskill
- , Phil Buckley
- , Anda Bularga
- , Ed Bullmore
- , Jenny Bunker
- , L. Burden
- , Tracy Burdett
- , David Burn
- , John Busby
- , Robyn Butcher
- , Al-Tahoor Butt
- , P. Cairns
- , P. C. Calder
- , Ellen Calvelo
- , H. Carborn
- , Bethany Card
- , Caitlin Carr
- , Liesel Carr
- , G. Carson
- , Penny Carter
- , Anna Casey
- , M. Cassar
- , Jonathon Cavanagh
- , Manish Chablani
- , Trudie Chalder
- , James D. Chalmers
- , Rachel Chambers
- , Flora Chan
- , K. M. Channon
- , Kerry Chapman
- , Amanda Charalambou
- , N. Chaudhuri
- , A. Checkley
- , Yutung Cheng
- , Luke Chetham
- , Caroline Childs
- , Edwin Chilvers
- , H. Chinoy
- , A. Chiribiri
- , K. Chong-James
- , N. Choudhury
- , Gaunab Choudhury
- , Phillip Chowienczyk
- , C. Christie
- , Melanie Chrystal
- , Cameron Clark
- , David Clark
- , Jude Clarke
- , S. Clohisey
- , G. Coakley
- , Zach Coburn
- , S. Coetzee
- , Joby Cole
- , Chris Coleman
- , Florence Conneh
- , David Connell
- , Bronwen Connolly
- , Lynda Connor
- , Amanda Cook
- , Shirley Cooper
- , B. Cooper
- , Josh Cooper
- , Donna Copeland
- , Tracey Cosier
- , Eamon Coughlan
- , Martina Coulding
- , C. Coupland
- , Thelma Craig
- , Daniele Cristiano
- , Michael Crooks
- , Andy Cross
- , Isabel Cruz
- , P. Cullinan
- , D. Cuthbertson
- , Luke Daines
- , Matthhew Dalton
- , Patrick Daly
- , Alison Daniels
- , Joanne Dasgin
- , Anthony David
- , Ffyon Davies
- , Ellie Davies
- , Kim Davies
- , Gareth Davies
- , Gwyneth Davies
- , Melanie Davies
- , Joy Dawson
- , Camilla Dawson
- , Enya Daynes
- , Anthony De Soyza
- , Bill Deakin
- , Andrew Deans
- , Joanne Deery
- , Sylviane Defres
- , Amanda Dell
- , K. Dempsey
- , Emma Denneny
- , J. Dennis
- , Ruvini Dharmagunawardena
- , Nawar Diar-Bakerly
- , Caroline Dickens
- , A. Dipper
- , Sarah Diver
- , Shalin Diwanji
- , Myles Dixon
- , R. Djukanovic
- , Hannah Dobson
- , S. L. Dobson
- , Annemarie B. Docherty
- , A. Donaldson
- , N. Dormand
- , Andrew Dougherty
- , Rachael Dowling
- , Stephen Drain
- , Katharine Draxlbauer
- , Katie Drury
- , Pearl Dulawan
- , A. Dunleavy
- , Sarah Dunn
- , Catherine Dupont
- , Joanne Earley
- , Nicholas Easom
- , Carlos Echevarria
- , Sarah Edwards
- , C. Edwardson
- , Claudia Efstathiou
- , Anne Elliott
- , K. Elliott
- , Yvette Ellis
- , Anne Elmer
- , Omer Elneima
- , Hosni El-Taweel
- , Teriann Evans
- , Ranuromanana Evans
- , Rachael A. Evans
- , Jonathon Evans
- , Cerys Evenden
- , Lynsey Evison
- , Laura Fabbri
- , Sara Fairbairn
- , Alexandra Fairman
- , K. Fallon
- , David Faluyi
- , Clair Favager
- , Tamanah Fayzan
- , James Featherstone
- , T. Felton
- , V. Ferreira
- , Selina Finney
- , J. Finnigan
- , L. Finnigan
- , Helen Fisher
- , S. Fletcher
- , Rachel Flockton
- , Margaret Flynn
- , David Foote
- , Amber Ford
- , D. Forton
- , Eva Fraile
- , C. Francis
- , Richard Francis
- , Susan Francis
- , Anew Frankel
- , Emily Fraser
- , N. French
- , Jonathon Fuld
- , J. Furniss
- , Lucie Garner
- , N. Gautam
- , John Geddes
- , J. George
- , P. George
- , Michael Gibbons
- , Rhyan Gill
- , Mandy Gill
- , L. Gilmour
- , F. Gleeson
- , Jodie Glossop
- , Sarah Glover
- , Nicola Goodman
- , Camelia Goodwin
- , Bibek Gooptu
- , Hussain Gordon
- , T. Gorsuch
- , M. Greatorex
- , Paul Greenhaff
- , William Greenhalf
- , Alan Greenhalgh
- , Neil J. Greening
- , John Greenwood
- , Rebecca Gregory
- , Heidi Gregory
- , D. Grieve
- , Denise Griffin
- , L. Griffiths
- , Anne-Marie Guerdette
- , Beatriz Guillen-Guio
- , Mahitha Gummadi
- , Ayushman Gupta
- , Sambasivarao Gurram
- , Elspeth Guthrie
- , Kate Hadley
- , Ahmed Haggar
- , Kera Hainey
- , Brigid Hairsine
- , Pranab Haldar
- , Lucy Hall
- , Mark Halling-Brown
- , Alyson Hancock
- , Kia Hancock
- , Neil Hanley
- , Sulaimaan Haq
- , Hayley Hardwick
- , Tim Hardy
- , Beverley Hargadon
- , Kate Harrington
- , Edward Harris
- , Victoria C. Harris
- , Ewen Harrison
- , Paul Harrison
- , Nicholas Hart
- , Alice Harvey
- , Matt Harvey
- , M. Harvie
- , L. Haslam
- , Claire Hastie
- , May Havinden-Williams
- , Jenny Hawkes
- , Nancy Hawkings
- , Jill Haworth
- , A. Hayday
- , Matthew Haynes
- , J. Hazeldine
- , Tracy Hazelton
- , Liam Heaney
- , Cheryl Heeley
- , Jonathon Heeney
- , M. Heightman
- , Simon Heller
- , Max Henderson
- , Helen Henson
- , L. Hesselden
- , Melanie Hewitt
- , Victoria Highett
- , T. Hillman
- , Ling-Pei Ho
- , Michaela Hoare
- , Amy Hoare
- , J. Hockridge
- , Philip Hogarth
- , Ailsa Holbourn
- , Sophie Holden
- , L. Holdsworth
- , D. Holgate
- , Maureen Holland
- , Leah Holloway
- , Katie Holmes
- , Megan Holmes
- , B. Holroyd-Hind
- , Anil Hormis
- , Alexander Horsley
- , Akram Hosseini
- , M. Hotopf
- , Linzy Houchen-Wolloff
- , Luke S. Howard
- , Kate Howard
- , Alice Howell
- , E. Hufton
- , Rachel Ann Hughes
- , Joan Hughes
- , Alun Hughes
- , Amy Humphries
- , Nathan Huneke
- , E. Hurditch
- , John Hurst
- , Masud Husain
- , Tracy Hussell
- , John Hutchinson
- , W. Ibrahim
- , Julie Ingham
- , L. Ingram
- , Diana Ionita
- , Karen Isaacs
- , Khalida Ismail
- , T. Jackson
- , Joseph Jacob
- , W. Y. James
- , Claire Jarman
- , Ian Jarrold
- , Hannah Jarvis
- , Roman Jastrub
- , Bhagy Jayaraman
- , Gisli Jenkins
- , P. Jezzard
- , Kasim Jiwa
- , C. Johnson
- , Simon Johnson
- , Desmond Johnston
- , Caroline Jolley
- , Ian Jones
- , Heather Jones
- , Mark Jones
- , Don Jones
- , Sherly Jose
- , Thomas Kabir
- , G. Kaltsakas
- , Vicky Kamwa
- , N. Kanellakis
- , Sabina Kaprowska
- , Zunaira Kausar
- , Natalie Keenan
- , Steven Kerr
- , Helen Kerslake
- , Angela Key
- , Fasih Khan
- , Kamlesh Khunti
- , Susan Kilroy
- , Bernie King
- , Clara King
- , Lucy Kingham
- , Jill Kirk
- , Paaig Kitterick
- , Paul Klenerman
- , Lucy Knibbs
- , Sean Knight
- , Abigail Knighton
- , Onn Min Kon
- , Samantha Kon
- , Ania Korszun
- , Ivan Koychev
- , Claire Kurasz
- , Prathiba Kurupati
- , Hanan Lamlum
- , G. Landers
- , Claudia Langenberg
- , Lara Lavelle-Langham
- , Allan Lawrie
- , Cathy Lawson
- , Claire Lawson
- , Alison Layton
- , Olivia C. Leavy
- , Ju Hee Lee
- , Elvina Lee
- , Karen Leitch
- , Rebecca Lenagh
- , Victoria Lewis
- , Joanne Lewis
- , Keir Lewis
- , N. Lewis-Burke
- , Felicity Liew
- , Tessa Light
- , Liz Lightstone
- , W. Lilaonitkul
- , S. Linford
- , Anne Lingford-Hughes
- , M. Lipman
- , Kamal Liyanage
- , Arwel Lloyd
- , Nazir I. Lone
- , Ronda Loosley
- , Janet Lord
- , Harpreet Lota
- , Wayne Lovegrove
- , Daniel Lozano-Rojas
- , Alice Lucey
- , Gardiner Lucy
- , E. Lukaschuk
- , Alison Lye
- , Ceri Lynch
- , S. MacDonald
- , G. MacGowan
- , Irene Macharia
- , J. Mackie
- , L. Macliver
- , S. Madathil
- , Gladys Madzamba
- , Nick Magee
- , Murphy Magtoto
- , N. Majeed
- , Flora Malein
- , Georgia Mallison
- , William Man
- , S. Mandal
- , K. Mangion
- , C. Manisty
- , R. Manley
- , Katherine March
- , Stefan Marciniak
- , Philip Marino
- , Myril Mariveles
- , Michael Marks
- , Elizabeth Marouzet
- , Sophie Marsh
- , M. Marshall
- , B. Marshall
- , Jane Martin
- , Adrian Martineau
- , L. M. Martinez
- , Nick Maskell
- , Darwin Matila
- , Wadzanai Matimba-Mupaya
- , Laura Matthews
- , Angeline Mbuyisa
- , Steve McAdoo
- , Hamish McAllister-Williams
- , Paul McArdle
- , Anne McArdle
- , Danny McAulay
- , Hamish J. C. McAuley
- , Gerry McCann
- , W. McCormick
- , Jacqueline McCormick
- , P. McCourt
- , Celeste McCracken
- , Lorcan McGarvey
- , Jade McGinness
- , K. McGlynn
- , Andrew McGovern
- , Heather McGuinness
- , I. B. McInnes
- , Jerome McIntosh
- , Emma McIvor
- , Katherine McIvor
- , Laura McLeavey
- , Aisling McMahon
- , Michael McMahon
- , L. McMorrow
- , Teresa Mcnally
- , M. McNarry
- , J. McNeill
- , Alison McQueen
- , H. McShane
- , Chloe Mears
- , Clare Megson
- , Sharon Megson
- , J. Meiring
- , Lucy Melling
- , Mark Mencias
- , Daniel Menzies
- , Marta Merida Morillas
- , Alice Michael
- , Benedict Michael
- , C. A. Miller
- , Lea Milligan
- , Nicholas Mills
- , Clare Mills
- , George Mills
- , L. Milner
- , Jane Mitchell
- , Abdelrahman Mohamed
- , Noura Mohamed
- , S. Mohammed
- , Philip Molyneaux
- , Will Monteiro
- , Silvia Moriera
- , Anna Morley
- , Leigh Morrison
- , Richard Morriss
- , A. Morrow
- , Paul Moss
- , Alistair Moss
- , K. Motohashi
- , N. Msimanga
- , Elizabeta Mukaetova-Ladinska
- , Unber Munawar
- , Jennifer Murira
- , Uttam Nanda
- , Heeah Nassa
- , Mariam Nasseri
- , Rashmita Nathu
- , Aoife Neal
- , Robert Needham
- , Paula Neill
- , Stefan Neubauer
- , D. E. Newby
- , Helen Newell
- , J. Newman
- , Tom Newman
- , Alex Newton-Cox
- , T. E. Nichols
- , Tim Nicholson
- , Christos Nicolaou
- , Debby Nicoll
- , Athanasios Nikolaidis
- , C. Nikolaidou
- , C. M. Nolan
- , Matthew Noonan
- , C. Norman
- , Petr Novotny
- , Kimon Ntotsis
- , Jose Nunag
- , Lorenza Nwafor
- , Uchechi Nwanguma
- , Joseph Nyaboko
- , Linda O’Brien
- , C. O’Brien
- , Natasha Odell
- , Kate O’Donnell
- , Godwin Ogbole
- , Olaoluwa Olaosebikan
- , Catherine Oliver
- , Zohra Omar
- , Peter J. M. Openshaw
- , D. P. O’Regan
- , Lorna Orriss-Dib
- , Lynn Osborne
- , Rebecca Osbourne
- , Marlies Ostermann
- , Charlotte Overton
- , Jamie Pack
- , Edmund Pacpaco
- , Stella-Maria Paddick
- , Sharon Painter
- , Erola Pairo-Castineira
- , Ashkan Pakzad
- , Sue Palmer
- , Padmasayee Papineni
- , K. Paques
- , Kerry Paradowski
- , Manish Pareek
- , Dhruv Parekh
- , H. Parfrey
- , Carmen Pariante
- , S. Parker
- , M. Parkes
- , J. Parmar
- , Sheetal Patale
- , Manish Patel
- , Suhani Patel
- , Dibya Pattenadk
- , M. Pavlides
- , Sheila Payne
- , Lorraine Pearce
- , John Pearl
- , Dan Peckham
- , Jessica Pendlebury
- , Yanchun Peng
- , Chris Pennington
- , Ida Peralta
- , Emma Perkins
- , Z. Peterkin
- , Tunde Peto
- , Nayia Petousi
- , John Petrie
- , Paul Pfeffer
- , Janet Phipps
- , S. Piechnik
- , John Pimm
- , Karen Piper Hanley
- , Riinu Pius
- , Hannah Plant
- , Tatiana Plekhanova
- , Megan Plowright
- , Krisnah Poinasamy
- , Oliver Polgar
- , Julie Porter
- , Joanna Porter
- , Sofiya Portukhay
- , Natassia Powell
- , A. Prabhu
- , James Pratt
- , Andrea Price
- , Claire Price
- , Carly Price
- , Anne Prickett
- , I. Propescu
- , J. Propescu
- , Sabrina Prosper
- , S. Pugmire
- , Sheena Quaid
- , Jackie Quigley
- , Jennifer K. Quint
- , H. Qureshi
- , I. N. Qureshi
- , K. Radhakrishnan
- , Najib Rahman
- , Markus Ralser
- , Betty Raman
- , Hazel Ramos
- , Albert Ramos
- , Jade Rangeley
- , Bojidar Rangelov
- , Liz Ratcliffe
- , Phillip Ravencroft
- , Konrad Rawlik
- , Anne Reddington
- , Heidi Redfearn
- , Dawn Redwood
- , Annabel Reed
- , Meryl Rees
- , Tabitha Rees
- , Karen Regan
- , Will Reynolds
- , Carla Ribeiro
- , A. Richards
- , Emma Richardson
- , M. Richardson
- , Pilar Rivera-Ortega
- , K. Roberts
- , Elizabeth Robertson
- , Leanne Robinson
- , Emma Robinson
- , Lisa Roche
- , C. Roddis
- , J. Rodger
- , Natalie Rogers
- , Gavin Ross
- , Alexandra Ross
- , Jennifer Rossdale
- , Anthony Rostron
- , Anna Rowe
- , J. Rowland
- , M. J. Rowland
- , A. Rowland
- , Sarah L. Rowland-Jones
- , Maura Roy
- , Igor Rudan
- , Richard Russell
- , Emily Russell
- , Gwen Saalmink
- , Ramsey Sabit
- , Beth Sage
- , T. Samakomva
- , Nilesh Samani
- , A. A. Samat
- , Claire Sampson
- , Katherine Samuel
- , Reena Samuel
- , Z. B. Sanders
- , Amy Sanderson
- , Elizabeth Sapey
- , Dinesh Saralaya
- , Jack Sargant
- , Carol Sarginson
- , Naveed Sattar
- , Kathryn Saunders
- , Peter Saunders
- , Ruth Saunders
- , Laura Saunders
- , Heather Savill
- , Avan Sayer
- , J. Schronce
- , William Schwaeble
- , Janet Scott
- , Kathryn Scott
- , Nick Selby
- , Malcolm G. Semple
- , Marco Sereno
- , Terri Ann Sewell
- , Kamini Shah
- , Ajay Shah
- , Manu Shankar-Hari
- , M. Sharma
- , Claire Sharpe
- , Michael Sharpe
- , Sharlene Shashaa
- , Alison Shaw
- , Victoria Shaw
- , Karen Shaw
- , Aziz Sheikh
- , Sarah Shelton
- , Liz Shenton
- , K. Shevket
- , Aarti Shikotra
- , Sulman Siddique
- , Salman Siddiqui
- , J. Sidebottom
- , Louise Sigfrid
- , Gemma Simons
- , Neil Simpson
- , John Simpson
- , Ananga Singapuri
- , Suver Singh
- , Claire Singh
- , Sally Singh
- , D. Sissons
- , J. Skeemer
- , Katie Slack
- , David Smith
- , Nikki Smith
- , Andrew Smith
- , Jacqui Smith
- , Laurie Smith
- , Susan Smith
- , M. Soares
- , Teresa Solano
- , Reanne Solly
- , A. R. Solstice
- , Tracy Soulsby
- , David Southern
- , D. Sowter
- , Mark Spears
- , Lisa Spencer
- , Fabio Speranza
- , Louise Stadon
- , Stefan Stanel
- , R. Steeds
- , N. Steele
- , Mike Steiner
- , David Stensel
- , G. Stephens
- , Lorraine Stephenson
- , Iain Stewart
- , R. Stimpson
- , Sue Stockdale
- , J. Stockley
- , Wendy Stoker
- , Roisin Stone
- , Will Storrar
- , Andrew Storrie
- , Kim Storton
- , E. Stringer
- , Sophia Strong-Sheldrake
- , Natalie Stroud
- , Christian Subbe
- , Catherine Sudlow
- , Zehra Suleiman
- , Charlotte Summers
- , C. Summersgill
- , Debbie Sutherland
- , D. L. Sykes
- , Nick Talbot
- , Ai Lyn Tan
- , Lawrence Tarusan
- , Vera Tavoukjian
- , Jessica Taylor
- , Abigail Taylor
- , Chris Taylor
- , John Paul Taylor
- , Amelie Te
- , Caroline Tee
- , J. Teixeira
- , Helen Tench
- , Sarah Terry
- , Susannah Thackray-Nocera
- , Favas Thaivalappil
- , David Thickett
- , David Thomas
- , S. Thomas
- , Caradog Thomas
- , Andrew Thomas
- , T. Thomas-Woods
- , A. A. Roger Thompson
- , Tamika Thompson
- , T. Thornton
- , Matthew Thorpe
- , Ryan S. Thwaites
- , Jo Tilley
- , N. Tinker
- , Gerlynn Tiongson
- , Martin Tobin
- , Johanne Tomlinson
- , Mark Toshner
- , T. Treibel
- , K. A. Tripp
- , Drupad Trivedi
- , E. M. Tunnicliffe
- , Alison Turnbull
- , Kim Turner
- , Sarah Turner
- , Victoria Turner
- , E. Turner
- , Sharon Turney
- , Lance Turtle
- , Helena Turton
- , Jacinta Ugoji
- , R. Ugwuoke
- , Rachel Upthegrove
- , Jonathon Valabhji
- , Maximina Ventura
- , Joanne Vere
- , Carinna Vickers
- , Ben Vinson
- , Ioannis Vogiatzis
- , Elaine Wade
- , Phillip Wade
- , Louise V. Wain
- , Tania Wainwright
- , Lilian Wajero
- , Sinead Walder
- , Samantha Walker
- , S. Walker
- , Tim Wallis
- , Sarah Walmsley
- , Simon Walsh
- , J. A. Walsh
- , Louise Warburton
- , T. J. C. Ward
- , Katie Warwick
- , Helen Wassall
- , Samuel Waterson
- , L. Watson
- , Ekaterina Watson
- , James Watson
- , M. Webster
- , J. Weir McCall
- , Carly Welch
- , Simon Wessely
- , Sophie West
- , Heather Weston
- , Helen Wheeler
- , Sonia White
- , Victoria Whitehead
- , J. Whitney
- , S. Whittaker
- , Beverley Whittam
- , V. Whitworth
- , Andrew Wight
- , James Wild
- , Martin Wilkins
- , Dan Wilkinson
- , Nick Williams
- , N. Williams
- , B. Williams
- , Jenny Williams
- , S. A. Williams-Howard
- , Michelle Willicombe
- , Gemma Willis
- , James Willoughby
- , Ann Wilson
- , Imogen Wilson
- , Daisy Wilson
- , Nicola Window
- , M. Witham
- , Rebecca Wolf-Roberts
- , Chloe Wood
- , F. Woodhead
- , Janet Woods
- , Dan Wootton
- , J. Wormleighton
- , J. Worsley
- , David Wraith
- , Caroline Wrey Brown
- , C. Wright
- , S. Wright
- , Louise Wright
- , Inez Wynter
- , Moucheng Xu
- , Najira Yasmin
- , S. Yasmin
- , Tom Yates
- , Kay Por Yip
- , Susan Young
- , Bob Young
- , A. J. Yousuf
- , Amira Zawia
- , Lisa Zeidan
- , Bang Zhao
- , Bang Zheng
- & O. Zongo
- , Daniel Agranoff
- , Ken Agwuh
- , Katie A. Ahmed
- , Dhiraj Ail
- , Erin L. Aldera
- , Ana Alegria
- , Beatrice Alex
- , Sam Allen
- , Petros Andrikopoulos
- , Brian Angus
- , Jane A. Armstrong
- , Abdul Ashish
- , Milton Ashworth
- , Innocent G. Asiimwe
- , Dougal Atkinson
- , Benjamin Bach
- , Siddharth Bakshi
- , Wendy S. Barclay
- , Shahedal Bari
- , Gavin Barlow
- , Samantha L. Barlow
- , Stella Barnass
- , Nicholas Barrett
- , Christopher Bassford
- , Sneha Basude
- , David Baxter
- , Michael Beadsworth
- , Jolanta Bernatoniene
- , John Berridge
- , Nicola Best
- , Debby Bogaert
- , Laura Booth
- , Pieter Bothma
- , Benjamin Brennan
- , Robin Brittain-Long
- , Katie Bullock
- , Naomi Bulteel
- , Tom Burden
- , Andrew Burtenshaw
- , Nicola Carlucci
- , Gail Carson
- , Vikki Caruth
- , Emily Cass
- , Benjamin W. A. Catterall
- , David Chadwick
- , Duncan Chambler
- , Meera Chand
- , Kanta Chechi
- , Nigel Chee
- , Jenny Child
- , Srikanth Chukkambotla
- , Richard Clark
- , Tom Clark
- , Jordan J. Clark
- , Emily A. Clarke
- , Sara Clohisey
- , Sarah Cole
- , Paul Collini
- , Marie Connor
- , Graham S. Cooke
- , Louise Cooper
- , Catherine Cosgrove
- , Audrey Coutts
- , Helen Cox
- , Jason Cupitt
- , Maria-Teresa Cutino-Moguel
- , Ana da Silva Filipe
- , Jo Dalton
- , Paul Dark
- , Christopher Davis
- , Chris Dawson
- , Thushan de Silva
- , Samir Dervisevic
- , Oslem Dincarslan
- , Alejandra Doce Carracedo
- , Cara Donegan
- , Lorna Donelly
- , Phil Donnison
- , Chloe Donohue
- , Gonçalo dos Santos Correia
- , Sam Douthwaite
- , Thomas M. Drake
- , Andrew Drummond
- , Marc-Emmanuel Dumas
- , Chris Dunn
- , Jake Dunning
- , Ingrid DuRand
- , Ahilanadan Dushianthan
- , Tristan Dyer
- , Philip Dyer
- , Angela Elliott
- , Cariad Evans
- , Anthony Evans
- , Chi Eziefula
- , Cameron J. Fairfield
- , Angie Fawkes
- , Chrisopher Fegan
- , Lorna Finch
- , Adam Finn
- , Lewis W. S. Fisher
- , Lisa Flaherty
- , Tom Fletcher
- , Terry Foster
- , Duncan Fullerton
- , Carrol Gamble
- , Isabel Garcia-Dorival
- , Atul Garg
- , Sanjeev Garg
- , Tammy Gilchrist
- , Michelle Girvan
- , Effrossyni Gkrania-Klotsas
- , Jo Godden
- , Arthur Goldsmith
- , Clive Graham
- , Tassos Grammatikopoulos
- , Christopher A. Green
- , Julian Griffin
- , Fiona Griffiths
- , Philip Gunning
- , Rishi K. Gupta
- , Katarzyna Hafezi
- , Sophie Halpin
- , Elaine Hardy
- , Ewen M. Harrison
- , Janet Harrison
- , Catherine Hartley
- , Stuart Hartshorn
- , Daniel Harvey
- , Peter Havalda
- , Daniel B. Hawcutt
- , Ross Hendry
- , Antonia Y. W. Ho
- , Maria Hobrok
- , Luke Hodgson
- , Karl Holden
- , Anthony Holmes
- , Peter W. Horby
- , Joanne Howard
- , Samreen Ijaz
- , Clare Jackson
- , Michael Jacobs
- , Susan Jain
- , Paul Jennings
- , Rebecca L. Jensen
- , Christopher B. Jones
- , Trevor R. Jones
- , Agilan Kaliappan
- , Vidya Kasipandian
- , Seán Keating
- , Stephen Kegg
- , Michael Kelsey
- , Jason Kendall
- , Caroline Kerrison
- , Ian Kerslake
- , Shadia Khandaker
- , Katharine King
- , Robyn T. Kiy
- , Stephen R. Knight
- , Susan Knight
- , Oliver Koch
- , Gouri Koduri
- , George Koshy
- , Chrysa Koukorava
- , Shondipon Laha
- , Eva Lahnsteiner
- , Steven Laird
- , Annette Lake
- , Suzannah Lant
- , Susan Larkin
- , Diane Latawiec
- , Andrew Law
- , James Lee
- , Gary Leeming
- , Daniella Lefteri
- , Tamas Leiner
- , Lauren Lett
- , Matthew Lewis
- , Sonia Liggi
- , Patrick Lillie
- , Wei Shen Lim
- , James Limb
- , Vanessa Linnett
- , Jeff Little
- , Lucia A. Livoti
- , Mark Lyttle
- , Louise MacGillivray
- , Alan Maclean
- , Michael MacMahon
- , Emily MacNaughton
- , Maria Mancini
- , Ravish Mankregod
- , Laura Marsh
- , Lynn Maslen
- , Hannah Massey
- , Huw Masson
- , Elijah Matovu
- , Nicole Maziere
- , Sarah McCafferty
- , Katherine McCullough
- , Sarah E. McDonald
- , Sarah McDonald
- , Laurence McEvoy
- , Ruth McEwen
- , John McLauchlan
- , Kenneth A. Mclean
- , Manjula Meda
- , Alexander J. Mentzer
- , Laura Merson
- , Soeren Metelmann
- , Alison M. Meynert
- , Nahida S. Miah
- , Joanna Middleton
- , Gary Mills
- , Jane Minton
- , Joyce Mitchell
- , Kavya Mohandas
- , James Moon
- , Elinoor Moore
- , Shona C. Moore
- , Patrick Morgan
- , Kirstie Morrice
- , Craig Morris
- , Katherine Mortimore
- , Samuel Moses
- , Mbiye Mpenge
- , Rohinton Mulla
- , Derek Murphy
- , Lee Murphy
- , Michael Murphy
- , Ellen G. Murphy
- , Thapas Nagarajan
- , Megan Nagel
- , Mark Nelson
- , Lisa Norman
- , Lillian Norris
- , Lucy Norris
- , Mahdad Noursadeghi
- , Michael Olanipekun
- , Wilna Oosthuyzen
- , Anthonia Osagie
- , Matthew K. O’Shea
- , Igor Otahal
- , Mark Pais
- , Massimo Palmarini
- , Carlo Palmieri
- , Selva Panchatsharam
- , Danai Papakonstantinou
- , Hassan Paraiso
- , Brij Patel
- , Natalie Pattison
- , William A. Paxton
- , Rebekah Penrice-Randal
- , Justin Pepperell
- , Mark Peters
- , Mandeep Phull
- , Jack Pilgrim
- , Stefania Pintus
- , Tim Planche
- , Daniel Plotkin
- , Georgios Pollakis
- , Frank Post
- , Nicholas Price
- , David Price
- , Tessa Prince
- , Rachel Prout
- , Nikolas Rae
- , Andrew Rambaut
- , Henrik Reschreiter
- , Tim Reynolds
- , Neil Richardson
- , P. Matthew Ridley
- , Mark Roberts
- , Stephanie Roberts
- , Devender Roberts
- , David L. Robertson
- , Alistair Rose
- , Guy Rousseau
- , Bobby Ruge
- , Clark D. Russell
- , Brendan Ryan
- , Debby Sales
- , Taranprit Saluja
- , Vanessa Sancho-Shimizu
- , Caroline Sands
- , Egle Saviciute
- , Matthias Schmid
- , Janet T. Scott
- , James Scott-Brown
- , Aarti Shah
- , Prad Shanmuga
- , Anil Sharma
- , Catherine A. Shaw
- , Victoria E. Shaw
- , Anna Shawcross
- , Rebecca K. Shears
- , Jagtur Singh Pooni
- , Jeremy Sizer
- , Benjamin Small
- , Richard Smith
- , Catherine Snelson
- , Tom Solomon
- , Rebecca G. Spencer
- , Nick Spittle
- , Shiranee Sriskandan
- , Nikki Staines
- , Tom Stambach
- , Richard Stewart
- , David Stuart
- , Krishanthi S. Subramaniam
- , Pradeep Subudhi
- , Olivia V. Swann
- , Tamas Szakmany
- , Agnieska Szemiel
- , Aislynn Taggart
- , Sarah Tait
- , Zoltan Takats
- , Panteleimon Takis
- , Jolanta Tanianis-Hughes
- , Kate Tatham
- , Richard S. Tedder
- , Jo Thomas
- , Jordan Thomas
- , Robert Thompson
- , Chris Thompson
- , Emma C. Thomson
- , Ascanio Tridente
- , Erwan Trochu
- , Darell Tupper-Carey
- , Lance C. W. Turtle
- , Mary Twagira
- , Nick Vallotton
- , Libby van Tonder
- , Rama Vancheeswaran
- , Rachel Vincent
- , Lisa Vincent-Smith
- , Shico Visuvanathan
- , Alan Vuylsteke
- , Sam Waddy
- , Rachel Wake
- , Andrew Walden
- , Ingeborg Welters
- , Murray Wham
- , Tony Whitehouse
- , Paul Whittaker
- , Ashley Whittington
- , Meme Wijesinghe
- , Eve Wilcock
- , Martin Williams
- , Lawrence Wilson
- , Stephen Winchester
- , Martin Wiselka
- , Adam Wolverson
- , Daniel G. Wootton
- , Andrew Workman
- , Nicola Wrobel
- , Bryan Yates
- , Peter Young
- , Maria Zambon
- & J. Eunice Zhang
Contributions
F.L. recruited participants, acquired clinical samples, analyzed and interpreted data and cowrote the manuscript, including all drafting and revisions. C.E. analyzed and interpreted data and cowrote this manuscript, including all drafting and revisions. S.F. and M.R. supported the analysis and interpretation of data as well as drafting and revisions. D.S., J.K.S., S.C.M., S.A., N.M., J.N., C.K., O.C.L., O.E., H.J.C.M., A. Shikotra, A. Singapuri, M.S., V.C.H., M.T., N.J.G., N.I.L. and C.C. contributed to acquisition of data underlying this study. L.H.-W., A.A.R.T., S.L.R.-J., L.S.H., O.M.K., D.G.W., T.I.d.S. and A. Ho made substantial contributions to conception/design and implementation of this work and/or acquisition of clinical samples for this work. They have supported drafting and revisions of the manuscript. E.M.H., J.K.Q. and A.B.D. made substantial contributions to the study design as well as data access, linkage and analysis. They have supported drafting and revisions of this work. J.D.C., L.-P.H., A. Horsley, B.R., K.P., M.M. and W.G. made substantial contributions to the conception and design of this work and have supported drafting and revisions of this work. J.K.B. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, has made substantial contributions to conception and design of this work and has supported drafting and revisions of this work. M.G.S. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, sponsor/protocol chief investigator, has made substantial contributions to conception and design of this work and has supported drafting and revisions of this work. R.A.E. and L.V.W. are co-leads of PHOSP-COVID, made substantial contributions to conception and design of this work, the acquisition and analysis of data, and have supported drafting and revisions of this work. C.B. is the chief investigator of PHOSP-COVID and has made substantial contributions to conception and design of this work. R.S.T. and L.T. made substantial contributions to the acquisition, analysis and interpretation of the data underlying this study and have contributed to drafting and revisions of this work. P.J.M.O. obtained funding for ISARIC4C, is ISARIC4C consortium co-lead, sponsor/protocol chief investigator and has made substantial contributions to conception and design of this work. R.S.T. and P.J.M.O. have also made key contributions to interpretation of data and have co-written this manuscript. All authors have read and approve the final version to be published. All authors agree to accountability for all aspects of this work. All investigators within ISARIC4C and the PHOSP-COVID consortia have made substantial contributions to the conception or design of this study and/or acquisition of data for this study. The full list of authors within these groups is available in Supplementary Information .
Corresponding authors
Correspondence to Ryan S. Thwaites or Peter J. M. Openshaw .
Ethics declarations
Competing interests.
F.L., C.E., D.S., J.K.S., S.C.M., C.D., C.K., N.M., L.N., E.M.H., A.B.D., J.K.Q., L.-P.H., K.P., L.S.H., O.M.K., S.F., T.I.d.S., D.G.W., R.S.T. and J.K.B. have no conflicts of interest. A.A.R.T. receives speaker fees and support to attend meetings from Janssen Pharmaceuticals. S.L.R.-J. is on the data safety monitoring board for Bexero trial in HIV+ adults in Kenya. J.D.C. is the deputy chief editor of the European Respiratory Journal and receives consulting fees from AstraZeneca, Boehringer Ingelheim, Chiesi, GSK, Insmed, Janssen, Novartis, Pfizer and Zambon. A. Horsley is deputy chair of NIHR Translational Research Collaboration (unpaid role). B.R. receives honoraria from Axcella therapeutics. R.A.E. is co-lead of PHOSP-COVID and receives fees from AstraZenaca/Evidera for consultancy on LC and from AstraZenaca for consultancy on digital health. R.A.E. has received speaker fees from Boehringer in June 2021 and has held a role as European Respiratory Society Assembly 01.02 Pulmonary Rehabilitation secretary. R.A.E. is on the American Thoracic Society Pulmonary Rehabilitation Assembly program committee. L.V.W. also receives funding from Orion pharma and GSK and holds contracts with Genentech and AstraZenaca. L.V.W. has received consulting fees from Galapagos and Boehringer, is on the data advisory board for Galapagos and is Associate Editor for the European Respiratory Journal . A. Ho is a member of NIHR Urgent Public Health Group (June 2020–March 2021). M.M. is an applicant on the PHOSP study funded by NIHR/DHSC. M.G.S. acts as an independent external and nonremunerated member of Pfizer’s External Data Monitoring Committee for their mRNA vaccine program(s), is Chair of Infectious Disease Scientific Advisory Board of Integrum Scientific LLC, and is director of MedEx Solutions Ltd. and majority owner of MedEx Solutions Ltd. and minority owner of Integrum Scientific LLC. M.G.S.’s institution has been in receipt of gifts from Chiesi Farmaceutici S.p.A. of Clinical Trial Investigational Medicinal Product without encumbrance and distribution of same to trial sites. M.G.S. is a nonrenumerated member of HMG UK New Emerging Respiratory Virus Threats Advisory Group and has previously been a nonrenumerated member of the Scientific Advisory Group for Emergencies (SAGE). C.B. has received consulting fees and/or grants from GSK, AstraZeneca, Genentech, Roche, Novartis, Sanofi, Regeneron, Chiesi, Mologic and 4DPharma. L.T. has received consulting fees from MHRA, AstraZeneca and Synairgen and speakers’ fees from Eisai Ltd., and support for conference attendance from AstraZeneca. L.T. has a patent pending with ZikaVac. P.J.M.O. reports grants from the EU Innovative Medicines Initiative 2 Joint Undertaking during the submitted work; grants from UK Medical Research Council, GSK, Wellcome Trust, EU Innovative Medicines Initiative, UK National Institute for Health Research and UK Research and Innovation–Department for Business, Energy and Industrial Strategy; and personal fees from Pfizer, Janssen and Seqirus, outside the submitted work.
Peer review
Peer review information.
Nature Immunology thanks Ziyad Al-Aly and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Ioana Staicu was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended data fig. 1 penalized logistic regression performance..
Graphs show classification error and Area under curve (AUC) from the 50 repeats tenfold nested cross-validation used to optimise and assess the performance of PLR testing associations with each LC outcome relative to Recovered (n = 233): Cardio_Resp (n = 398), Fatigue (n = 384), Anxiety/Depression (n = 202), GI (n = 132), ( e ) Cognitive (n = 6). The distributions of classification error and area under curve (AUC) from the nested cross-validation are shown. Box plot centre line represents the Median and boundaries of the box represent interquartile range (IQR), the whisker length represent 1.5xIQR.
Extended Data Fig. 2 Associations with long COVID symptoms in full study cohort.
( a ) Fibrinogen levels at 6 months were compared between pooled LC cases (n = 295) and Recovered (n = 233) and between the Cognitive group (n = 41) and Recovered (n = 233). Box plot centre line represent the Median and boundaries of the box represent interquartile range (IQR), the whisker length represents 1.5xIQR, any outliers beyond the whisker range are shown as individual dots. Median differences were compared using two-sided Wilcoxon signed-rank test *= p < 0·05, **= p < 0·01, ***= p < 0·001, ****= p < 0·0001. Unadjusted p-values are reported. b ) Distribution of time from COVID-19 hospitalisation at sample collection applying CDC and NICE definitions of LC (n = 719) ( c ) Upset plot of symptom groups. Horizontal coloured bars represent the number of patients in each symptom group: Cardiorespiratory (Cardio_Resp), Fatigue, Cognitive, Gastrointestinal (GI) and Anxiety/Depression (Anx_Dep). Vertical black bars represent the number of patients in each symptom combination group. To prevent patient identification, where less than 5 patients belong to a combination group, this has been represented as ‘<5’. The Recovered group (n = 250) were used as controls. Forest plots show Olink protein concentrations (NPX) associated with ( d ) Cardio_Resp (n = 398), ( e ) Fatigue (n = 342), ( f ) Anx_Dep (n = 219), ( g ) GI (n = 134), and ( h ) Cognitive (n = 65). Error bars represent the median accuracy of the model.
Extended Data Fig. 3 Validation of olink measurements using conventional assays in plasma.
Olink measured protein (NPX) were compared to chemiluminescence assays (ECL or ELISA, log2[pg/mL]) to validate our findings, where contemporaneously collected plasma samples were available (n = 58). Results from key mediators associated with LC groups were validated: CSF3, IL1R2, IL2, IL3RA, TNFa, TFF2. R = spearman rank correlation coefficient and shaded areas indicated the 95% confidence interval. Samples that fell below the lower limit of detection for a given assay were excluded and the ‘n’ value on each panel indicates the number of samples above this limit.
Extended Data Fig. 4 Univariate analysis of proteins associated with each symptom.
Olink measured plasma protein levels (NPX) compared between LC groups (Cardio_Resp, n = 398, Fatigue n = 384, Anxiety/Depression, n = 202, GI, n = 132 and Cognitive, n = 60) and Recovered (n = 233). Proteins identified by PLR were compared between groups. Median differences were compared using two-sided Wilcoxon signed-rank test. * = p < 0·05, ** = p < 0·01, *** = p < 0·001, ****= p < 0·0001 after FDR adjustment. Box plot centre line represent the Median and boundaries of the box represent interquartile range (IQR), the whisker length represents 1.5xIQR, any outliers beyond the whisker range are shown as individual dots.
Extended Data Fig. 5 Unadjusted Penalised Logistic Regression.
Olink measured proteins (NPX) and their association with Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65). Forest plots show odds of each LC outcome vs Recovered (n = 233), using PLR without adjusting for clinical co-variates. Error bars represent the median accuracy of the model.
Extended Data Fig. 6 Partial Least Squares analysis.
Olink measured proteins (NPX) and their association with Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups. Forest plots show odds of LC outcome vs Recovered (n = 233), using PLS analysis. Error bars represent the standard error of the coefficient estimate.
Extended Data Fig. 7 Network analysis centrality.
Each graph shows the centrality score for each Olink measured protein (NPX) found to have significant associations with other proteins that were elevated in the Cardio_Resp (n = 398), Fatigue (n = 342), Anx_Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups relative to Recovered (n = 233).
Extended Data Fig. 8 Inflammation in men and women with long COVID.
Olink measured plasma protein levels (NPX) between men and women with symptoms, divided by age (<50 or >=50years): (a) shows IL1R2 and MATN2 in the Anxiety/Depression group (<50 n = 55, >=50 n = 133), (b) shows CTSO and NFASC in the Cognitive group (<50 n = 11, >=50 n = 50). Median values were compared between men and women using two-sided Wilcoxon signed-rank test. Box plot centre line represent the Median and boundaries represent interquartile range (IQR), the whisker length represents 1.5xIQR.
Extended Data Fig. 9 Inflammation in the upper respiratory tract.
Nasal cytokines measured by immunoassay in the CardioResp Group (n = 29) and Recovered (n = 31): ( a ) shows IL1a, IL1b, IL-6, APO-2, TGFa, TFF2. Median differences were compared using two-sided Wilcoxon signed-rank test. Box plot centre line represents the Median and boundaries of the box represent interquartile range (IQR), the whisker length represent 1.5xIQR. ( b ) Shows cytokines measured by immunoassay in paired plasma and nasal (n = 70). Correlations between IL1a, IL1b, IL-6, APO-2, TGFa and TFF2 in nasal and plasma samples were compared using Spearman’s rank correlation coefficient ( R ). Shaded areas indicated the 95% confidence interval of R.
Extended Data Fig. 10 Graphical abstract.
Summary of interpretation of key findings from Olink measured proteins and their association with CardioResp (n = 398), Fatigue (n = 342), Anx/Dep (n = 219), GI (n = 134), and Cognitive (n = 65) groups relative to Recovered (n = 233).
Supplementary information
Supplementary information.
Supplementary Methods, Statistics and reproducibility statement, Supplementary Results, Supplementary Tables 1–7, Extended data figure legends, Appendix 1 (Supplementary Table 8), Appendix 2 (PHOSP-COVID author list) and Appendix 3 (ISARIC4C author list).
Reporting Summary
Rights and permissions.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
Liew, F., Efstathiou, C., Fontanella, S. et al. Large-scale phenotyping of patients with long COVID post-hospitalization reveals mechanistic subtypes of disease. Nat Immunol 25 , 607–621 (2024). https://doi.org/10.1038/s41590-024-01778-0
Download citation
Received : 11 August 2023
Accepted : 06 February 2024
Published : 08 April 2024
Issue Date : April 2024
DOI : https://doi.org/10.1038/s41590-024-01778-0
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
Immune dysregulation in long covid.
- Laura Ceglarek
- Onur Boyman
Nature Immunology (2024)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Open access
- Published: 02 April 2024
Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus : a review
- Junqing Qiao 1 ,
- Rainer Borriss 2 ,
- Kai Sun 3 ,
- Rongsheng Zhang 1 ,
- Xijun Chen 3 ,
- Youzhou Liu 1 &
- Yongfeng Liu 1
Microbial Cell Factories volume 23 , Article number: 100 ( 2024 ) Cite this article
252 Accesses
2 Altmetric
Metrics details
Surfactin is a cyclic hexalipopeptide compound, nonribosomal synthesized by representatives of the Bacillus subtilis species complex which includes B. subtilis group and its closely related species, such as B. subtilis subsp subtilis, B. subtilis subsp spizizenii, B. subtilis subsp inaquosorum , B. atrophaeus, B. amyloliquefaciens, B. velezensis (Steinke mSystems 6: e00057, 2021) It functions as a biosurfactant and signaling molecule and has antibacterial, antiviral, antitumor, and plant disease resistance properties. The Bacillus lipopeptides play an important role in agriculture, oil recovery, cosmetics, food processing and pharmaceuticals, but the natural yield of surfactin synthesized by Bacillus is low. This paper reviews the regulatory pathways and mechanisms that affect surfactin synthesis and release, highlighting the regulatory genes involved in the transcription of the srfAA-AD operon. The several ways to enhance surfactin production, such as governing expression of the genes involved in synthesis and regulation of surfactin synthesis and transport, removal of competitive pathways, optimization of media, and fermentation conditions were commented. This review will provide a theoretical platform for the systematic genetic modification of high-yielding strains of surfactin.
Graphical Abstract
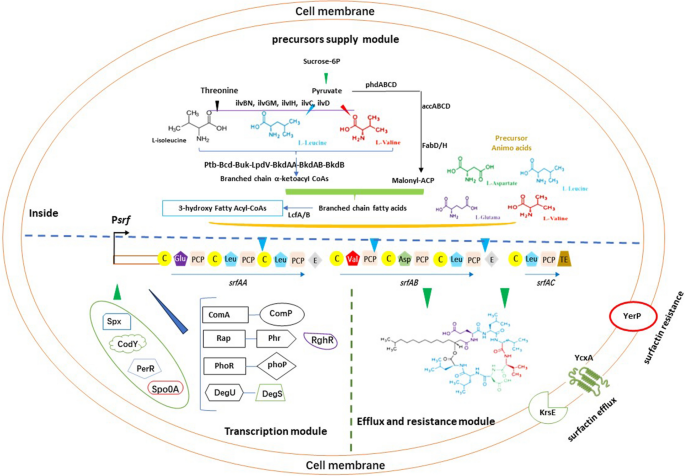
Introduction
Recently, biopesticides made by antagonistic Bacillus species or their metabolites have been used to reduce the application of chemical pesticides in agriculture [ 1 ]. The most used Bacillus species include Bacillus subtilis, Bacillus velezensis, and Bacillus thuringiensis , such as B. velezensis FZB42, B. subtilis Bs916, B. subtilis QST713, and B. thuringiensis HD1, which were used in rice, wheat, maize, cotton, tomato, lettuce and cucumber [ 2 , 3 , 4 , 5 ]. The biocontrol efficiency of Bacillus strain relies on three main traits: ecological fitness, strong antagonistic activity toward plant pathogens, and an ability to trigger plant immune reaction [ 6 ]. The cyclic lipopeptides (LPs) of the surfactin, iturin and fengycin families produced by Bacillus strains are the key biocontrol elements which contribute to the traits mentioned above, especially antagonistic activity of biocontrol agents [ 6 , 7 , 8 ]. Especially surfactin is a key element in plant-bacteria interactions, enabling the Bacillus cells to form biofilms on plant roots, and to stimulate induced systemic resistance in plants. Cross talks between plants and bacteria are accomplished by a bacterial sensing system for plant pectin leading to enhanced surfactin synthesis in B. velezensis [ 9 ]. In addition, surfactin is directly or indirectly involved in several processes of cell differentiation, such as development of competence, motility, matrix production, cannibalism, quorum sensing and endospore formation [ 10 , 11 , 12 ].
Surfactin is a representative biosurfactant of the cyclic lipopeptide family which is synthesized by nonribosomal peptide synthetases (NRPS) and then transported outside the cells. Its chemical structure consists of a cycloheptapeptide, which is interlinked with β-hydroxy fatty acid chains of variable length containing 12−17 carbon atoms. Giant biosynthetic gene clusters (BGCs) involved in synthesis of surfactin were detected in many representatives of the Bacillus subtilis species complex, such as B. subtilis , B. atrophaeus , B. spizizenii , B. amyloliquefaciens , and B. velezensis [ 1 , 13 ]. The surfactin BGC (e. g. BGC0000433, B. velezensis FZB42 , [ 14 ] contains a co-encoded regulatory gene comS , which is located within the open reading frame of the srfAB gene [ 15 ]. ComS is involved in regulation of genetic competence [ 16 ], and simultaneously part of the quorum sensing system [ 17 ]. Natural variants of the surfactant surfactin are lichenysin from B. licheniformis [ 18 ] and pumilacidin from B. pumilus [ 19 ].
Due to its complex unique structure, surfactin can reduce the surface tension of water from 72 to 27.90 mN /m and possesses high thermal stability and salt resistance. Surfactin is also among the most well-known lipopeptide antibiotics with broad-spectrum antibacterial, antiviral and antitumor properties. Surfactin has great potential applications in agriculture, oil recovery, cosmetics, food processing and pharmaceuticals because of its structural stability, surfactant and antibiotic activity [ 20 , 21 , 22 ]. However, surfactin commercial application has been hindered due to the low yield obtained from Bacillus cultures. Therefore, several investigations have been focused on identification of the key regulatory mechanisms of surfactin biosynthesis to enhance surfactin production.
This review outlines a general overview of the regulation mechanisms affecting surfactin biosynthesis, and comments the attempts to enhance surfactin production. Starting with the regulation of transcription of the srfAA-AD operon, we also review the mechanisms of the export of surfactin from the cells. Special attention is given to the genes involved in synthesis of the necessary precursors for surfactin synthesis, such as branched-chain fatty acids, and amino acids, which are available only in limited amounts in the producer cells. Other factors, suitable to enhance yield of surfactin, such as removal of competitive metabolic pathways, and optimization of media and fermentation conditions were also considered. This review is aimed to provide a knowledge base for applying systematic genetic and other strategies for enhancing surfactin production, and for generating novel surfactin variants.
Genes affecting surfactin expression
To increase the production of surfactin, several scientists have investigated the regulatory pathways and mechanisms that affect its synthesis and release. On the transcriptional level, expression of the surfactin biosynthesis pathway is—besides their general control—mainly influenced by five features: (1) the supply of β-hydroxy fatty acids; (2) the biosynthesis of α-amino acids; (3) the assembly of fatty-acid chains and amino acids, in which amino acids are sequentially assembled onto fatty acyl-coenzyme A through the NRPS system; (4) the release of surfactin from the Bacillus cells; (5) removal of other gene clusters involved in competitive synthesis pathways. The mechanism of secretion of surfactin is not fully understood, and excess surfactin can be toxic to Bacillus cells [ 23 ]. The gene categories mentioned above directly or indirectly regulate the synthesis and release of surfactin. The specific regulatory genes are listed in Table 1
A multitude of regulators affect transcription of the srfAA-AD operon
The srfAA-AD operon transcribes the nonribosomal peptide synthetases and a thioesterase involved in surfactin synthesis (Fig. 1 ). The NRPS system consists of seven catalytic modules encoded by the genes srfAA , srfAB , srfAC , which is 27 kb in length [ 67 , 68 ]. Each catalytic module is responsible for the recognition and condensation of an amino acid, and each module contains several catalytic structural domains: an adenylation (a) structural domain; a peptidyl carrier protein or thiolation (PCP) structural domain; a condensation (C) structural domain; a differential isomerization (E) domain; and a thioesterase (TE) structural domain, which is only present in the termination module [ 69 , 70 , 71 ]. Module 1 is responsible for the condensation of l-Glu; modules 2, 3, 6, and 7 are responsible for the condensation of l-Leu; module 4 is responsible for the condensation of l-Val; and module 5 is responsible for the condensation of l-Asp. srfAD is a type II thioesterase gene responsible for the cyclization and release of surfactin [ 72 ]. Downstream to the srfAA-AD operon, there is a phosphopantothionine ethantransferase (PPTases) gene, encoding the Sfp protein that activates surfactin synthesis [ 73 ]. Deletion of the sfp gene results in the inability to synthesize all three classes of lipopeptide antibiotics in Bacillus [ 74 ].
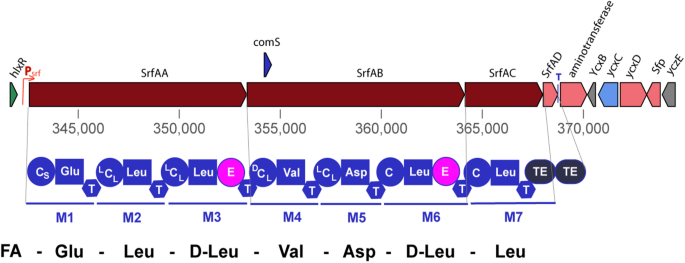
The surfactin gene cluster (BGC0000433) in B. velezensis FZB42. Transcription of the srfAA-AD operon is governed by the P srf promoter. The comS gene is embedded within the srfAB gene, and is also transcribed by the P srf promoter. Three genes, srfAA , srfAB , and srfAC , transcribe the nonribosomal peptide synthetases (NRPS) SrfAA, SrfAB, SrfAC, and the thioesterase SrfAD. SrfAA contains N-terminally the CS-domain and acylates the first amino acid Glu1 with various fatty acids. The elongation modules of SrfAA, SrfAB, and SrfAC yield the linear heptapeptide indicated at the bottom of the figure. The TE domain in SrfAC releases the lipopeptide and performs the cyclization between Leu7, and the fatty acid chain linked with Glu1. The second TE domain present in SrfAD seems to have mainly repair functions. The sfp gene, located downstream from the srfAA-AD operon encodes a phosphopantetheinyl transferase (PPTase), which is indispensable for nonribosomal synthesis of surfactin, the other lipopeptides (fengycin and bacillomycin D), and polyketides in FZB42. The yczE gene product, a membrane protein with unknown function, was also shown to be essential for synthesis of cyclic lipopeptides
The srfAA-AD operon transcription is governed by the SigA-dependent promoter P srf . Studies have shown that srfAA-AD expression is influenced by several regulatory factors and pathways (Fig. 2 ). Transcription of the srfAA-AD operon is activated by binding of phosphorylated ComA within the surfactin promoter region upstream of the srfAA gene. Further transcription factors, positively affecting transcription of the srfAA-AD operon are PerR and PhoP, whilst Abh, CodY, and Spx have a negative effect, and can be removed in order to enhance transcription of the surfactin operon.
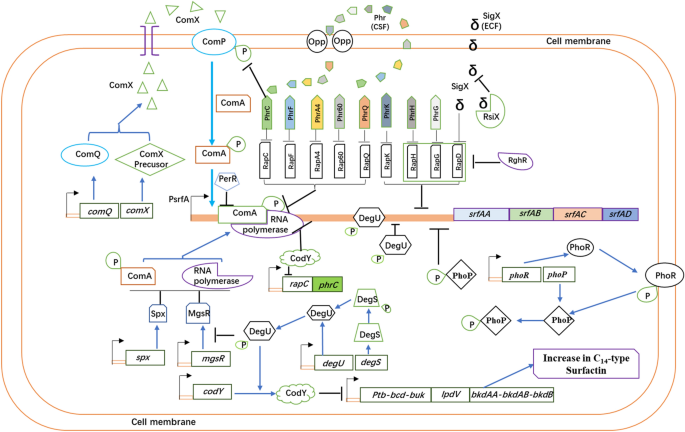
Schematic model for the regulation pathway of transcription of the srfAA-AD cluster. Black bent arrow represents the promoter of gene or operon. Black T-bar indicates the negative effects on DNA binding or protein interactions. ‘P’ in the circle means the phosphoryl group
The ComA-ComP two-component system
The ComA-ComP two-component system is the primary regulatory system governing transcription of the srfAA-AD operon. The synthesis of surfactin starts with the production of ComX, an extracellular peptide pheromone that is continuously synthesized, and accumulated during cell growth. When the cells reach a certain density, ComX is sensed, and undergoes autophosphorylation by the histidine kinase ComP, and subsequently interacts with ComA to phosphorylate ComA (ComA-P). ComA-P then activates the srfAA-AD operon transcription by binding at two binary symmetric regions upstream of the P srf promoter-35 sequence, and is therefore essential for transcription of the srfAA-AD operon [ 24 , 25 , 26 ].
The Rap-Phr two-component system
The Rap-Phr system, consisting of aspartate phosphatases (Rap) and their inhibitory oligopeptides (Phr), governs fine tuning of the ComA-P dependent transcription of the surfactin operon during cell growth [ 36 ]. Six Rap proteins (C, D, F, G, H, and K) are involved. RapC and RapF suppress srfAA-AD expression by binding with ComA, which competes with the ComA phosphorylation exerted by ComP [ 31 , 32 ]. RapD, RapG, RapH, and RapK overexpression also leads to inhibition of the srfAA-AD transcription. Site-directed mutagenesis has shown that rapG disruption increases srfAA-AD expression at least twofold, whilst rapH disruption has little effect on srfAA-AD expression [ 33 , 35 , 36 ].
Rap protein activity is inhibited by the Phr peptides. For survival in hostile environments, Bacillus species utilizes the Rap-Phr two component system to govern the differentiation of its populations, where the Phr pentapeptides function as quorum sensing signals [ 75 , 76 ]. The phr gene is activated during the transition phase between exponential growth and stationary phase to express a precursor peptide with a putative signal peptide. After export of the pre-Phr from the cell via the SecA secretion system, the signal peptide is cleaved off, and the active Phr pentapeptide is generated [ 77 ]. The Phr peptides are imported into the cell by the oligopeptide permease (Opp) system, and act as quorum sensing signal, when the population density reached a high level. Then, inside the cell, they inhibit Rap protein activity [ 78 , 79 ]. B. subtilis encodes five Phr peptides (PhrC, PhrF, PhrG, PhrH, and PhrK) that inhibit the activity of their cognate Rap proteins, and so enable the Bacillus cells to overcome the Rap-dependent inhibition of the surfactin expression [ 34 , 35 ]. Studies have demonstrated that the extracellular signaling molecule PhrC enters the cell and binds to the RapC protein, which inhibits the interaction between RapC and ComA-P and promotes the subsequent transcription of the srfAA-AD gene [ 28 , 30 ]. Jung et al. [ 27 ] successfully increased srfAA-AD gene transcription and surfactin production by overexpressing the comX and phrC genes in B. subtilis pHT43. Liang et al. [ 29 ] demonstrated that a novel RapA4-PhrA4 system of B. amyloliquefaciens NAU-B3 regulates surfactin production similar to RapC-PhrC. RapD lacks the homologous Phr peptide. As such, RapD is positively regulated by the extracellular function of the σ-factor SigX, whereas SigX is negatively regulated by its cognate anti-σ factor RsiX. The disruption of rsiX results in the accumulation of SigX, which increases RapD production. Elevated levels of RapD downregulate the srfAA-AD expression in B. subtilis cells [ 40 ]. Furthermore, RghR can repress rapD , rapG , and rapH expression in B. subtilis cells. RghR indirectly increases srfAA-AD transcription by specifically binding to the promoter sequences of rapD , rapG , and rapH , which suppresses rapD , rapG , and rapH expression [ 33 , 35 ].
The Rap60-Phr60 system found in the B. subtilis endogenous plasmid pTA1060 regulates surfactin expression. Rap60 regulates ComA activity in a manner that is unique to B. subtilis 168 by forming a ternary complex with ComA and DNA. The resulting complex inhibits ComA activity without interfering with DNA binding. In comparison, reactions involving RapC demonstrate alterations in DNA binding [ 37 ]. Phr60 is an important inhibitor of Rap60 activity. It was also demonstrated that the RapQ-PhrQ system of the endogenous cryptic plasmid pBSG3 in B. amyloliquefaciens B3 regulates surfactin production, competence and sporulation similar to Rap60–Phr60 system [ 38 , 39 ].
The PhoR-PhoP two-component system
The PhoR-PhoP two-component system positively regulates surfactin production under phosphate limitation [ 44 ]. Dong et al. [ 43 ] demonstrated that srfAA gene expression was reduced in PhoR and PhoP mutants in low phosphorus conditions. Wild-type strain NCD-2 produced 2.3–6.4 times more surfactin than the PhoR and PhoP mutant strains. In a low phosphorus environment, the histidine protein kinase PhoR autophosphorylates PhoR-P, which is located on the cell membrane. It then transfers a phosphate group to the response regulator PhoP, located in the cytoplasm. PhoP-P regulates target gene expression by binding to the promoter region of the downstream target gene [ 41 , 42 ].
The DegU-DegS two-component system
The two-component regulatory system DegU-DegS is involved in the expression of extracellular protease, and lipopeptide antibiotics during late growth phase [ 80 ]. DegU owning typical helix-turn-helix DNA sequence-binding structures, has an ability to regulate the gene transcription by binding to the promoter regions through phosphorylated DegU (DegU-P) or unphosphorylated DegU [ 81 ]. DegU regulates bacillomycin D biosynthesis positively, and surfactin biosynthesis negatively [ 45 , 46 , 47 , 48 ]. In 2016, Mathieu et al. [ 46 ] reported that the DegU-DegS system differentially regulates the “K-state” which is a growth-arrested state of cells induced by ComK regulon [ 82 ] in undomesticated wild and laboratory domesticated B. subtilis model strains. This, in turn, affects the efficiency of competence cell formation. The specific regulatory mechanisms associated with this interaction are as follows: a site-specific mutation in the degQ promoter of domesticated laboratory strain reduces degQ expression capacity. Lower degQ expression lowers DegU-P concentration within the cell; however, this has little or no effect on the transcription of the surfactin operon. Moreover, the ComS levels remain stable, which, in turn, increases the levels of the receptor transcriptional regulator ComK. Bacteria are more likely to enter the K-state, and to form receptor cells. Unmodified wild-type Bacillus cells have no point mutation in the degQ promoter. As such, these cells have high intracellular concentrations of DegU-P, which inhibit srfAA-AD operon transcription. Deletion of the degQ gene in B. subtilis led to a threefold increase in surfactin production [ 83 ]. These cells also have low levels of ComS, which decrease ComK levels. The bacteria are then less likely to enter the K-state, and less able to form competent cells. Research has also shown that unphosphorylated DegU can activate the transcription of ComK. Our recent study showed that degU mutation resulted in a significant increase of surfactin and decrease of fengycin in wild-type strain B. subtilis Bs916. It is possible that DegU directly binds to the srfAA promoter region or indirectly regulates srfAA-AD expression by regulating CodY, PhrC, and MgsR (unpublished).
The Spx, CodY, PerR and Spo0A proteins
The Spx, CodY and PerR regulator proteins in B. subtilis hinder the srfAA-AD promoter transcription. Spx prevents the ComA-P dependent srfAA-AD promoter transcription by occupying overlapping sites in the αCTD region of RNA polymerase. This inhibits the interaction between ComA-P and RNA polymerase on the srfAA promoter, which inhibits the expression of the srfAA-AD operon [ 51 , 52 ]. PerR represses also transcription by competitive binding on the ComA-P binding site at the srfAA promoter region [ 53 ]. When the spx and perR genes in B. subtilis are suppressed, srfAA-AD transcription levels increase by 4.5–4.2-fold, respectively [ 56 ].
The CodY protein in B. subtilis is a global regulator that inhibits srfAA-AD transcription by competing with the RNA polymerase binding sites in the srfAA promoter region [ 50 , 84 ]; High amino acid concentrations activate CodY, and enable its binding within the srfAA promoter region. Knockout of the codY gene in B. subtilis 168 increases surfactin production by approximately tenfold [ 62 ]. CodY also represses the transcription of the bkd gene cluster that is involved in branched-chain ketoacid and fatty acid biosynthesis [ 85 ], deletion of codY also results in changes of surfactin isoforms.
Phosphorylation of the master regulator Spo0A initiates the sporulation process by inhibiting AbrB leading to transcription of competence and sporulation factor CSF, and was shown to negatively control surfactin synthesis. Knock out of the spo0A gene enhances surfactin synthesis [ 49 ].
Genes associated with branched-chain fatty acid synthesis in surfactin
Fatty acids are key structural elements in surfactin. As such, fatty acid biosynthesis, particularly of branched-chain fatty acids, is essential for surfactin synthesis [ 63 , 64 ]. A large number of intermediates are involved in this biosynthetic pathway. Researchers have demonstrated that surfactin production is dependent on the regulation of certain intermediates.
Genes associated with Malonyl-coenzyme-A synthesis
The acetyl-CoA carboxylase (ACCase) enzyme complex is encoded by four genes ( accA , accB , accC , and accD ). The complex contains two key catalytic structural domains: a biotin carboxylase encoded by accC , and a carboxyltransferase encoded by accA and accD . Furthermore, accB participates in the reaction by encoding the biotin carboxyl carrier protein that attaches to the cofactor biotin [ 54 ]. B. subtilis catalyzes the formation of malonyl-CoA from acetyl-CoA through ACCase. This is the first and rate-limiting step in fatty acid synthesis. Wang et al. [ 55 ] demonstrated that an yngH -encoded ACCase subunit (biotin carboxylase II) could maintain acetyl-CoA ACCase activity. Inhibition of this particular ACCase subunit resulted in significantly greater decreases in ACCase activity and surfactin production. In contrast, overexpression of yngH in B. subtilis TS1726 significantly increased ACCase activity, and surfactin production increased by 43%. In addition, when accBC and accAD expression are blocked with antisense RNA, there is a small decrease in ACCase activity and surfactin production, respectively.
Genes associated with malonyl-acyl carrier protein (ACP) synthesis
Malonyl-CoA is converted to malonyl-ACP by acyl carrier protein transacylase (FabD). In B. subtilis 168, overexpression of accABCD and fabD increased surfactin production slightly by 14% [ 56 ].
Genes associated with β-keto acyl-ACP synthesis
The β-ketoacyl-acyl carrier protein synthase III (fabHB) catalyzes the condensation of β-keto acyl-ACP from malonyl-ACP and branched α-ketoacyl CoA. This reaction is the first step in branched-chain fatty acid biosynthesis. Wu et al. [ 56 ] enhanced branched-chain fatty acid synthesis by overexpressing fabHB in B. subtilis 168, which resulted in a significant increase in surfactin production.
Genes associated with 3-hydroxy fatty acyl-CoA synthesis
The final substrate involved in assembly of the surfactin fatty acid chain is 3-hydroxy fatty acyl-CoA. This molecule requires one of four fatty acyl-CoA ligases (LcfA, YhfL, YhfT, or YngI) [ 59 , 60 ]. When LcfA or YhfL are involved in the reaction, CoA thioesters are formed from the combination of 3-hydroxy fatty acids and CoA. These thioesters are recognized by the donor site of the C-structural domain of the first module, which catalyzes the nucleophilic attack of the α-amino group of the PCP-bound glutamate in the thioester bond of the fatty acid. The resulting acylated glutamate is accepted by the next C-structural domain, which allows peptide assembly to continue. Once complete, the final molecule is released [ 57 , 58 ]. In comparison, when YhfT is involved in the reaction, only acyl adenylate intermediates can be observed, and no CoA thioesters are formed. Instead, YhfT plays a role in surfactin production by transferring acyl-adenosine monophosphate (AMP) derivatives to ACP, which, in turn, transfers intermediates to the surfactin assembly line. Meanwhile, the role of YngI in surfactin production in B. subtilis requires further elucidation.
Kraas et al. [ 57 ] demonstrated that the knockout of lcfA , yhfL , yhfT or yngI in B. subtilis OKB105 reduced surfactin production by 38–55%. After knock out of all four genes surfactin production was found reduced by 84%. Since the complete deletion did not completely eliminate surfactin production, it can be assumed that other pathways provide fatty acids for surfactin production. This also shows that branched-chain fatty acid biosynthesis plays a significant role in surfactin biosynthesis.
Genes involved in biosynthesis of the amino acids used in surfactin synthesis
Besides fatty acids, amino acids are important precursors for surfactin biosynthesis. Increasing the amount of available amino acid precursors increases surfactin production. In addition, it was shown that the production of surfactin could be regulated by affecting the pathway of biosynthesis of four amino acids (l-Glu, l-Leu, l-Val and l-Asp). One study found that enhancing the leucine metabolic pathway resulted in a 20.9-fold increase in surfactin production [ 62 ]. Wang et al. [ 61 ] demonstrated that B. subtilis 168 increased surfactin production when genes yrpC , racE , or murC , which are associated with l-Glu consumption in the branching pathway, were repressed. In contrast, when genes pyrB or pyrC , which participate in the branching pathway for l-Asp biosynthesis, were suppressed, surfactin production was reduced.
The genes bkdAA and bkdAB are involved in pathways leading to consumption of l-Leu and l-Val. Suppression of bkdAA or bkdAB increases surfactin production and alters the proportion of final surfactin isoforms; For example, C 14 -type surfactin increases by almost 60% [ 63 ]. BkdAA and BkdAB initiate these reactions by increasing the accumulation of l-Leu and l-Val, and blocking the bkd operon ( lpdV , bkdAA , bkdAB , and bkdB ) which resulted in the reducing of iso-C 13 and iso-C 15 fatty acid synthesis [ 64 ]. Furthermore, Dhali et al. [ 63 ] demonstrated that disruption of the dehydrogenase complex in lpdV mutants inhibited branched-chain amino acid utilization and CoA precursor conversion into their respective branched-chain fatty acids. This resulted in a 2.5-fold increase in C 14 -type surfactin.
Branched-chain fatty acids and amino acids are the key structural elements of surfactin, and isomers with branched-chain fatty acid variants are the main components of surfactin variants, accounting for about 78% of the total. The branched-chain fatty acid synthesis precursors isobutyryl-CoA, isovaleryl-CoA and α-methylbutyryl-CoA originate from the branched-chain amino acids l-valine, l-leucine and l-isoleucine, respectively [ 21 ]. As described above, the biosynthesis of branched-chain fatty acids and amino acids significantly influenced the biosynthesis of surfactin. Therefore, it is possible to enhance the accumulation of branched fatty acid chains and amino acids for surfactin biosynthesis by modifying the metabolic pathways genetically, in order to improve the production of surfactin.
Overexpression of surfactin efflux pump genes avoid adverse action of excess surfactin
The above mentioned factors directly or indirectly affect surfactin synthesis, but the rate of surfactin release and the strain self-resistance to surfactin are important factors because excess surfactin concentrations are toxic to Bacillus cells. It was reported that surfactin began to destroy the integrity of the liposome (simulated cell membrane) at 10 mg/L, when the concertation of surfactin increased to 500 mg/L, the liposome thoroughly disappeared [ 66 ]. This indicated that the intracellular concentration of surfactin in the living microbial cells could not be too high, or the cell membrane might be destroyed. Tsuge et al. [ 23 ] reported that the cell survival rate of B. subtilis 168 was decreased with the increasing of surfactin. The cell survival rate was only 50% when the surfactin concentration in the medium reached 100 mg/L. Known lipopeptide transport proteins are YcxA, KrsE, and YerP, that utilize proton motive force as an energy source. Surfactin efflux is a two-step process wherein (1) the polar amino acid of surfactin reacts with certain amino acid residues at the substrate binding sites of YcxA or KrsE, and (2) the lipid fraction facilitates membrane binding and permeation through the hydrophobic channels formed by KrsE [ 65 ]. Li et al. [ 66 ] found that surfactin transfer was impossible in B. subtilis THY-7 strains bearing YcxA mutation, whilst overexpression of the full-length YcxA increased surfactin secretion by 89%. Overexpression of KrsE also increased surfactin production by 52%. YerP is homologous to the resistance and cytokinesis family of PMF-dependent efflux pumps. YerP has been shown to be essential for surfactin resistance in B. subtilis [ 23 ]. When YerP was overexpressed, surfactin production increased by 145%. Li et al. [ 66 ] postulated that YerP-mediated surfactin resistance is essentially similar to the transmembrane transport of surfactin by YerP. Taken together, overexpression of the genes acting as surfactin efflux pumps is a way to avoid cell toxicity caused by excess surfactin.
Approaches for enhancing surfactin production
Several studies have reported that the surfactin titer of wild-type Bacillus strains is limited to 100–600 mg/L [ 86 ]. Therefore, it seems difficult to achieve a break-through in the yield of surfactin only by traditional screening of wild strains, and by optimization of media and fermentation conditions. The systematic genetic modification, based on target directed changes in the complex regulatory network involved in surfactin expression, seems to be a promising strategy for constructing high-yielding surfactin producers. By combining systematic genetic modification with the use of novel and innovative fermentation methods, such as high-cell-density fermentation, a significant break-through of the surfactin yield can be achieved.
The probably most spectacular example for successful strain improvement based on metabolic engineering was achieved with the B. subtilis model strain 168, which is per se unable to synthesize surfactin due to a nonsense mutation in its sfp gene [ 56 ]. Therefore, the ability to produce surfactin was restored by integrating the wild-type sfp + gene, resulting in a surfactin yield of 0.4 g/L. Then, by removing of competing genes involved in biofilm formation, and nonribosomal synthesis of peptides and polyketides a 3.3-fold increase in productivity was obtained. In a further step, cellular resistance to surfactin was enhanced by overexpressing genes associated with export and self-resistance, such as swrC ( yerP ) and the liaIHGFSR operon. This results in an 8.5-fold increase of the surfactin titer. Next, by increased supply of branched-chain fatty acids, the surfactin yield was enhanced to 8.5 g/L corresponding to an increase of 20.3-fold. In a final step, supply of the available fatty acid precursor acetyl-CoA was enhanced by redirecting it from cell growth to surfactin synthesis. The final surfactin titer reached 12.8 g/L corresponding to 42% of the theoretical yield calculated for the substrate sucrose.
Applying alternative strategies, such as promoter substitution, genome reduction and genome shuffling might contribute also to higher surfactin yields [ 87 ]. Here, we shortly summarize the different strategies for enhancing surfactin production.
Enhancement of transcription of the surfactin operon
The regulatory network governing the ComA dependent transcription of the surfactin operon has been extensively discussed in Sect. A multitude of regulators affect transcription of the srfAA-AD operon . It is recommended to enhance the transcription of the srfAA-AD operon though overexpression of ComX, PhrC and ComA, and decreasing the amount of DegU, CodY, Spx, PerR, which leads to the accumulation of the nonribosomal peptide synthetases SrfAA, SrfAB, and SrfAC for surfactin biosynthesis and improves assembly of the peptide moiety. Furthermore, the knockdown of degU could significantly increase the production of surfactin, while the biosynthesis of iturin and fengycin was almost completely inhibited [ 45 ].
Promoter engineering
Surfactin expression can be enhanced by replacing the original P srf promoter with stronger promoters, such as P xyl , P spac , and P g3 [ 21 ]. Replacement of the natural P srf promoter by the IPTG inducible P spac promoter resulted in a tenfold increase of the surfactin yield [ 88 ]. By using the artificial P g3 promoter, the surfactin titer was enhanced from 0.55 g/L produced in the original B. subtilis THY-7–9.74 g/L produced in the engineered strain. 0.14 g surfactin were obtained per g sucrose [ 89 ].
Increase of the supply of the building blocks for surfactin synthesis
It is suggested that the metabolic pathways of fatty acids and amino acids could be genetically modified to increase the supply of branched fatty acid chains and the amino acids l-glutamate, l-leucine, l-valine, and l-aspartate involved in the surfactin biosynthesis. For instance, the suppression of the expression of the bkd operon ( lpdV, bkdAA, bkdAB and bkdB genes) not only increased the accumulation of l-Leu and l-Val, but also increased the iso-C 14 fatty acid accumulation [ 61 ]. By strengthening the leucine metabolic pathway, surfactin production was enhanced by more than 20-fold [ 62 ].
Enhanced export from the cell
Enhancing the efflux of surfactin avoids toxic effects to the cell due to high surfactin concentration, however the exact mechanisms of the transport of surfactin through the cell membrane are still not clear. It is assumed that transmembrane exporters dependent on proton motive force are involved. It was reported that the yerP gene is involved in surfactin self-resistance [ 23 ]. Overexpression of the putative surfactin transporters YcxA, KrsE, and SwrC (YerP) resulted in an enhanced secretion of surfactin [ 66 ].
Combinatorial biosynthesis for generation of more efficient surfactin
As well as surfactin high-yielding engineered strains, combinatorial biosynthesis strategies can also be used to modify the structure of surfactin to improve its effect. Combinatorial biosynthesis plays an important role in the structural modification of lipopeptides. It alters the lipopeptide biosynthetic pathway purposefully to create predictable structural products that exhibit new functions or activities as expected by the investigator. The structural modifications of the surfactin are mainly peptide rings and hydrophobic fatty acid chains [ 90 ].
Application of genome reduction and shuffling strategies
Different methods based on genomic rearrangement (“genome shuffling”) [ 91 ] and reduction of the genome size were applied to enhance surfactin production. Three rounds of genome shuffling via recursive protoplast fusion in B. velezensis resulted in a fourfold increase of productivity from 229.6 mg/L–908,15 mg/L [ 92 ].
A genome reduced B. subtilis strain in which 10% of the whole genome, including the fengycin and bacilysin gene clusters, was removed, was found superior in its growth parameters, but did not surpass the original strain in surfactin productivity [ 93 ].
Process and media optimization
The patent filed by Kaneka Corp. (US7011969B2) claims that surfactin concentrations of up to 50 g/L can be reached after a long-term fermentation of 80 h, performed with B. subtilis and soybean flour as carbon source. Unfortunately, a detailed description of the process parameters, and strain properties were not given [ 94 ].
High cell density fermentation following a fed-batch protocol was used for efficient surfactin production with the nonsporulating Bacillus subtilis strain 3NA, in which a sfp + gene has been introduced [ 49 ]. A cell density of 88 g/L accompanied with a surfactin titer of 26.5 g/L was reached after 38 h fermentation, impressively underlining the power of optimizing fermentation process parameters, together with the use of purposefully engineered high-yielding production strains.
Summary and outlook
In recent years, many efforts have been devoted to unravel the complex network involved in surfactin expression and to optimize the production process for a “green” surfactin to an economically attractive level. Rapid advances in the application of efficient genetic engineering techniques for the development of high-yielding strains, together with the use of high-cell density and other innovative fermentation techniques, now enable the production of surfactin in a range of 20 g/L and above. The success story reviewed here could also promote the development of highly efficient production processes for the biosynthesis of other “green” compounds which can be used as environmentally friendly tools in sustainable agriculture and industry. Examples include fengycins and iturin-like compounds with antifungal properties, as well as other active ingredients that are useful in biological plant protection.
Definitions of the key concepts
Nonribosomal peptide synthetases (NRPSs), a large multi-enzyme is composed of repeating enzyme domains with modular organization to activate and couple fatty acids to l-amino acids, l-amino acids to l-amino acids, and D-amino acids to l-amino acids in a particular order to generate linear or cyclic peptides.
Competence, a distinct DNA uptake phenotype of Bacillus subtilis which appears to be a cell survival strategy for either procuring new genetic information or obtaining DNA as food. The competence is correlated with high cell density and nutrient limiting conditions.
“K-state”, a growth-arrested state of Bacillus cells induced by transcription factor ComK, of which competence for genetic transformation is but one notable feature. This is a unique adaptation to stress and the persistent state has been defined the “K-state”.
Combinatorial biosynthesis, an approach to produce novel natural products with modifying of biosynthetic pathways by genetic engineering. The feasibility of this approach was demonstrated in biosynthesis of lipopeptide, polyketides and nucleoside antibiotics.
Genome shuffling, a method that combines DNA shuffling with the recombination of entire genomes which provide an alternative to the rapid production of improved strains in microorganisms metabolic engineering breeding.
Availability of data and materials
Not applicable.
Steinke K, Mohite OS, Weber T, Kovács Á. Phylogenetic distribution of secondary metabolites in the bacillus subtilis species complex. mSystems. 2021. https://doi.org/10.1128/mSystems.00057-21 .
Article PubMed PubMed Central Google Scholar
Borriss R. Use of plant-associated bacillus strains as biofertilizers and biocontrol agents in agriculture. In: Maheshwari D, editor. Bacteria in Agrobiology: Plant Growth Responses. Berlin, Heidelberg: Springer; 2011. p. 41–76.
Chapter Google Scholar
Mishra R, Arora AK, Jimenez J, Dos Santos TC, Banerjee R, Panneerselvam S, Bonning BC. Bacteria-derived pesticidal proteins active against hemipteran pests. J Invertebr Pathol. 2022;195: 107834.
Article CAS PubMed Google Scholar
Yamamoto T. Engineering of Bacillus thuringiensis insecticidal proteins. J Pestic Sci. 2022;47:47–58.
Article CAS PubMed PubMed Central Google Scholar
Yamamoto T. One hundred years of Bacillus thuringiensis research and development: discovery to transgenic crops. J Insect Biotechnol Sericolo. 2001;70:1–23.
CAS Google Scholar
Cawoy H, Debois D, Franzil L, De Pauw E, Thonart P, Ongena M. Lipopeptides as main ingredients for inhibition of fungal phytopathogens by Bacillus subtilis/amyloliquefaciens . Microb Biotechnol. 2015;8:281–95.
Seydlova G, Svobodova J. Review of surfactin chemical properties and the potential biomedical applications. Cent Eur J Med. 2008;3:123–33.
Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–25.
Hoff G, Arguelles Arias A, Boubsi F, Prsic J, Meyer T, Ibrahim HMM, Steels S, Luzuriaga P, Legras A, Franzil L, et al. Surfactin stimulated by pectin molecular patterns and root exudates acts as a key driver of the Bacillus –Plant mutualistic interaction. MBio. 2021;12: e0177421.
Article PubMed Google Scholar
Rahman FB, Sarkar B, Moni R, Rahman MS. Molecular genetics of surfactin and its effects on different sub-populations of Bacillus subtilis . Biotechnol Rep. 2021;32: e00686.
Article CAS Google Scholar
Chen B, Wen J, Zhao X, Ding J, Qi G. Surfactin: a quorum-sensing signal molecule to relieve CCR in Bacillus amyloliquefaciens . Front Microbiol. 2020;11:631.
Wen J, Zhao X, Si F, Qi G. Surfactin, a quorum sensing signal molecule, globally affects the carbon metabolism in. Metab Eng Commun. 2021;12: e00174.
Theatre A, Cano-Prieto C, Bartolini M, Laurin Y, Deleu M, Niehren J, Fida T, Gerbinet S, Alanjary M, Medema MH, et al. The surfactin-like lipopeptides from Bacillus spp.: natural biodiversity and synthetic biology for a broader application range. Front Bioeng Biotechnol. 2021. https://doi.org/10.3389/fbioe.2021.623701 .
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol. 2004;186:1084–96.
Hamoen LW, Eshuis H, Jongbloed J, Venema G, van Sinderen D. A small gene, designated comS , located within the coding region of the fourth amino acid activation domain of srfA , is required for competence development in Bacillus subtilis . Mol Microbiol. 1995;15:55–63.
Liu JJ, Zuber P. A molecular switch controlling competence and motility: competence regulatory factors ComS, MecA, and ComK control sigma(D)-dependent gene expression in Bacillus subtilis . J Bacteriol. 1998;180:4243–51.
Stiegelmeyer SM, Giddings MC. Agent-based modeling of competence phenotype switching in Bacillus subtilis . Theor Biol Med Model. 2013;10:23.
Horowitz S, Gilbert JN, Griffin WM. Isolation and characterization of a surfactant produced by Bacillus licheniformis 86. J Ind Microbiol. 1990;6:243–8.
Naruse N, Tenmyo O, Kobaru S, Kamei H, Miyaki T, Konishi M, Oki T. Pumilacidin, a complex of new antiviral antibiotics—production, isolation, chemical-properties structure and biological-activity. J Antibiot. 1990;43:267–80.
Bordoloi NK, Konwar BK. Microbial surfactant-enhanced mineral oil recovery under laboratory conditions. Colloids Surf B-Biointerfaces. 2008;63:73–82.
Hu F, Liu Y, Li S. Rational strain improvement for surfactin production: enhancing the yield and generating novel structures. Microb Cell Fact. 2019;18:42.
Kisil OV, Trefilov VS, Sadykova VS, Zvereva ME, Kubareva EA. Surfactin: its biological activity and possibility of application in agriculture. Appl Biochem Microbiol. 2023;59:1–13.
Tsuge K, Ohata Y, Shoda M. Gene yerP , involved in surfactin self-resistance in Bacillus subtilis . Antimicrob Agents Chemother. 2001;45:3566–73.
Bacon Schneider K, Palmer TM, Grossman AD. Characterization of comQ and comX , two genes required for production of ComX pheromone in Bacillus subtilis . J Bacteriol. 2002;184:410–9.
Karatas AY, Cetin S, Ozcengiz G. The effects of insertional mutations in comQ , comP , srfA , spo0H , spo0A and abrB genes on bacilysin biosynthesis in Bacillus subtilis . Biochim Biophys Acta. 2003;1626:51–6.
Roggiani M, Dubnau D. ComA, a phosphorylated response regulator protein of Bacillus subtilis , binds to the promoter region of srfA . J Bacteriol. 1993;175:3182–7.
Jung J, Yu KO, Ramzi AB, Choe SH, Kim SW, Han SO. Improvement of surfactin production in Bacillus subtilis using synthetic wastewater by overexpression of specific extracellular signaling peptides, comX and phrC . Biotechnol Bioeng. 2012;109:2349–56.
Lazazzera BA, Solomon JM, Grossman AD. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis . Cell. 1997;89:917–25.
Liang Z, Qiao JQ, Li PP, Zhang LL, Qiao ZX, Lin L, Yu CJ, Yang Y, Zubair M, Gu Q, et al. A novel Rap-Phr system in Bacillus velezensis NAU-B3 regulates surfactin production and sporulation via interaction with ComA. Appl Microbiol Biotechnol. 2020;104:10059–74.
Pottathil M, Jung A, Lazazzera BA. CSF, a species-specific extracellular signaling peptide for communication among strains of Bacillus subtilis and Bacillus mojavensis . J Bacteriol. 2008;190:4095–9.
Core L, Perego M. TPR-mediated interaction of RapC with ComA inhibits response regulator-DNA binding for competence development in Bacillus subtilis . Mol Microbiol. 2003;49:1509–22.
Bongiorni C, Ishikawa S, Stephenson S, Ogasawara N, Perego M. Synergistic regulation of competence development in Bacillus subtilis by two Rap-Phr systems. J Bacteriol. 2005;187:4353–61.
Hayashi K, Kensuke T, Kobayashi K, Ogasawara N, Ogura M. Bacillus subtilis RghR (YvaN) represses rapG and rapH , which encode inhibitors of expression of the srfA operon. Mol Microbiol. 2006;59:1714–29.
Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis . Nature. 1997;390:249–56.
Ogura M, Fujita Y. Bacillus subtilis rapD , a direct target of transcription repression by RghR, negatively regulates srfA expression. FEMS Microbiol Lett. 2007;268:73–80.
Auchtung JM, Lee CA, Grossman AD. Modulation of the ComA-dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J Bacteriol. 2006;188:5273–85.
Boguslawski KM, Hill PA, Griffith KL. Novel mechanisms of controlling the activities of the transcription factors Spo0A and ComA by the plasmid-encoded quorum sensing regulators Rap60-Phr60 in Bacillus subtilis . Mol Microbiol. 2015;96:325–48.
Qiao JQ, Tian DW, Huo R, Wu HJ, Gao XW. Functional analysis and application of the cryptic plasmid pBSG3 harboring the RapQ-PhrQ system in Bacillus amyloliquefaciens B3. Plasmid. 2011;65:141–9.
Yang Y, Wu HJ, Lin L, Zhu QQ, Borriss R, Gao XW. A plasmid-born Rap-Phr system regulates surfactin production, sporulation and genetic competence in the heterologous host, Bacillus subtilis OKB105. Appl Microbiol Biotechnol. 2015;99:7241–52.
Huang XJ, Helmann JD. Identification of target promoters for the Bacillus subtilis sigma(X) factor using a consensus-directed search. J Mol Biol. 1998;279:165–73.
Eldakak A, Hulett FM. Cys303 in the histidine kinase PhoR is crucial for the phosphotransfer reaction in the PhoPR two-component system in Bacillus subtilis . J Bacteriol. 2007;189:410–21.
Martin JF. Phosphate control of the biosynthesis of antibiotics and other secondary metabolites is mediated by the PhoR-PhoP system: an unfinished story. J Bacteriol. 2004;186:5197–201.
Dong LH, Guo QG, Wang PP, Li SZ, Lu XY, Zhang XY, W.S. Z, Ma P,. The effect of PhoR/PhoP two-component regulatory system on surfactin production in Bacillus subtilis NCD-2. Acta Phytopathologica Sinica. 2018;48:119–27.
Google Scholar
Salzberg LI, Botella E, Hokamp K, Antelmann H, Maass S, Becher D, Noone D, Devine KM. Genome-wide analysis of phosphorylated PhoP binding to chromosomal DNA reveals several novel features of the PhoPR-mediated phosphate limitation response in Bacillus subtilis . J Bacteriol. 2015;197:1492–506.
Koumoutsi A, Chen XH, Vater J, Borriss R. DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl Environ Microbiol. 2007;73:6953–64.
Miras M, Dubnau D. A DegU-P and DegQ-dependent regulatory pathway for the K-state in Bacillus subtilis . Front Microbiol. 1868;2016:7.
Sun J, Liu Y, Lin F, Lu Z, Lu Y. CodY, ComA, DegU and Spo0A controlling lipopeptides biosynthesis in Bacillus amyloliquefaciens fmbJ. J Appl Microbiol. 2021;131:1289–304.
Yu C, Qiao J, Ali Q, Jiang Q, Song Y, Zhu L, Gu Q, Borriss R, Dong S, Gao X, Wu H. degQ associated with the degS/degU two-component system regulates biofilm formation, antimicrobial metabolite production, and biocontrol activity in Bacillus velezensis DMW1. Mol Plant Pathol. 2023;24:1510–21.
Klausmann P, Hennemann K, Hoffmann M, Treinen C, Aschern M, Lilge L, Heravi KM, Henkel M, Hausmann R. Bacillus subtilis high cell density fermentation using a sporulation-Deficient strain for the production of surfactin. Appl Microbiol Biotechnol. 2021;105:4141–51.
Serror P, Sonenshein AL. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J Bacteriol. 1996;178:5910–5.
Newberry KJ, Nakano S, Zuber P, Brennan RG. Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci U S A. 2005;102:15839–44.
Zhang Y, Nakano S, Choi SY, Zuber P. Mutational analysis of the Bacillus subtilis RNA polymerase alpha C-terminal domain supports the interference model of Spx-dependent repression. J Bacteriol. 2006;188:4300–11.
Hayashi K, Ohsawa T, Kobayashi K, Ogasawara N, Ogura M. The H 2 O 2 stress-responsive regulator PerR positively regulates srfA expression in Bacillus subtilis . J Bacteriol. 2005;187:6659–67.
Wakil SJ, Stoops JK, Joshi VC. Fatty-acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–79.
Wang MM, Yu HM, Shen ZY. Antisense RNA-based strategy for enhancing surfactin production in Bacillus subtilis TS1726 via overexpression of the unconventional biotin Carboxylase II To enhance ACCase activity. ACS Synth Biol. 2019;8:251–6.
Wu Q, Zhi Y, Xu Y. Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis 168. Metab Eng. 2019;52:87–97.
Kraas FI, Helmetag V, Wittmann M, Strieker M, Marahiel MA. Functional dissection of surfactin synthetase initiation module reveals insights into the mechanism of lipoinitiation. Chem Biol. 2010;17:872–80.
Matsuoka H, Hirooka K, Fujita Y. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J Biol Chem. 2007;282:5180–94.
Menkhaus M, Ullrich C, Kluge B, Vater J, Vollenbroich D, Kamp RM. Structural and functional-organization of the surfactin synthetase multienzyme system. J Biol Chem. 1993;268:7678–84.
Steller S, Sokoll A, Wilde C, Bernhard F, Franke P, Vater J. Initiation of surfactin biosynthesis and the role of the SrfD-thioesterase protein. Biochemistry. 2004;43:11331–43.
Wang CY, Cao YX, Wang YP, Sun LM, Song H. Enhancing surfactin production by using systematic CRISPRi repression to screen amino acid biosynthesis genes in Bacillus subtilis . Microb Cell Fact. 2019;18:90.
Coutte F, Niehren J, Dhali D, John M, Versari C, Jacques P. Modeling leucine’s metabolic pathway and knockout prediction improving the production of surfactin, a biosurfactant from Bacillus subtilis . Biotechnol J. 2015;10:1216–34.
Dhali D, Coutte F, Arias AA, Auger S, Bidnenko V, Chataigne G, Lalk M, Niehren J, de Sousa J, Versari C, Jacques P. Genetic engineering of the branched fatty acid metabolic pathway of Bacillus subtilis for the overproduction of surfactin C 14 isoform. Biotechnol J. 2017;12:1600574.
Article Google Scholar
Kaneda T. Fatty acids of the genus Bacillus : an example of branched-chain preference. Bacteriol Rev. 1977;41:391–418.
Law CJ, Maloney PC, Wang DN. Ins and outs of major facilitator superfamily antiporters. Annu Rev Microbiol. 2008;62:289–305.
Li X, Yang H, Zhang D, Li X, Yu H, Shen Z. Overexpression of specific proton motive force dependent transporters facilitate the export of surfactin in Bacillus subtilis . J Ind Microbiol Biotechnol. 2015;42:93–103.
Nakano MM, Magnuson R, Myers A, Curry J, Grossman AD, Zuber P. srfA Is an operon required for surfactin production, competence development, and efficient sporulation in Bacillus subtilis . J Bacteriol. 1991;173:1770–8.
Nakano MM, Marahiel MA, Zuber P. Identification of a genetic-locus required for biosynthesis of the lipopeptide antibiotic surfactin in Bacillus subtilis . J Bacteriol. 1988;170:5662–8.
Durfahrt T, Marahiel MA. Peptide antibiotics from the molecular production line. Nachr Chem. 2005;53:507–13.
Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–73.
Mootz HD, Marahiel MA. Biosynthetic systems for nonribosomal peptide antibiotic assembly. Curr Opin Chem Biol. 1997;1:543–51.
Koglin A, Lohr F, Bernhard F, Rogov VV, Frueh DP, Strieter ER, Mofid MR, Guntert P, Wagner G, Walsh CT, et al. Structural basis for the selectivity of the external thioesterase of the surfactin synthetase. Nature. 2008;454:907–11.
Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998;37:1585–95.
Mootz HD, Finking R, Marahiel MA. 4 '-phosphopantetheine transfer in primary and secondary metabolism of Bacillus subtilis . J Biol Chem. 2001;276:37289–98.
D’Souza C, Nakano MM, Zuber P. Identification of comS , a gene of the srfA operon that regulates the establishment of genetic competence in Bacillus subtilis . Proc Natl Acad Sci U S A. 1994;91:9397–401.
Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis . Science. 2007;317:526–9.
Stephenson S, Mueller C, Jiang M, Perego M. Molecular analysis of Phr peptide processing in Bacillus subtilis . J Bacteriol. 2003;185:4861–71.
Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A. 1997;94:8612–7.
Tjalsma H, Bolhuis A, Jongbloed JD, Bron S, van Dijl JM. Signal peptide-dependent protein transport in Bacillus subtilis : a genome-based survey of the secretome. Mircobiol Mol Biol Rev. 2000;64:515–47.
Ogura M, Yamaguchi H, Ki Y, Fujita Y, Tanaka T. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B.subtilis two-component regulatory systems. Nucleic Acids Resea. 2001;29:3804–13.
Shimane K, Ogura M. Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters. J Biochem. 2004;136:387–97.
Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol Microbiol. 2002;43:1331–45.
Lilge L, Vahidinasab M, Adiek I, Becker P, Nesamani CK, Treinen C, Hoffmann M, Heravi KM, Henkel M, Hausmann R. Expression of degQ gene and its effect on lipopeptide production as well as formation of secretory proteases in Bacillus subtilis strains. MicrobiologyOpen. 2021;10: e1241.
Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–28.
Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of BkdR, a transcriptional activator of the SigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis . J Bacteriol. 1999;181:2059–66.
Liu XY, Ren BA, Chen M, Wang HB, Kokare CR, Zhou XL, Wang JD, Dai HQ, Song FH, Liu M, et al. Production and characterization of a group of bioemulsifiers from the marine Bacillus velezensis strain H3. Appl Microbiol Biotechnol. 2010;87:1881–93.
Xia L, Wen JP. Available strategies for improving the biosynthesis of surfactin: a review. Crit Rev Biotechnol. 2022;43:1111–28.
Sun HG, Bie XM, Lu FX, Lu YP, Wu YDL, Lu ZX. Enhancement of surfactin production of Bacillus subtilis fmbR by replacement of the native promoter with the Pspac promoter. Can J Microbiol. 2009;55:1003–6.
Jiao S, Li X, Yu HM, Yang H, Li X, Shen ZY. In situ enhancement of surfactin biosynthesis in Bacillus subtilis using novel artificial inducible promoters. Biotechnol Bioeng. 2017;114:832–42.
Baltz RH. Combinatorial biosynthesis of cyclic lipopeptide antibiotics: a model for synthetic biology to accelerate the evolution of secondary metabolite biosynthetic pathways. ACS Synth Biol. 2014;3:748–58.
Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WPC, del Cardayre SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–6.
Chen L, Chong XY, Zhang YY, Lv YY, Hu YS. Genome shuffling of Bacillus velezensis for enhanced surfactin production and variation analysis. Curr Microbiol. 2020;77:71–8.
Geissler M, Kuhle I, Heravi KM, Altenbuchner J, Henkel M, Hausmann R. Evaluation of surfactin synthesis in a genome reduced Bacillus subtilis strain. AMB Express. 2019;9:84–97.
YYM Tadashi, K Furuya, T Tsuzuki. Production process of surfactin. Kaneka Corp J (ed). Patent no. US7011969B2. 9. US2006: 9
Download references
Acknowledgements
We gratefully acknowledge the helpful discussions with Professor Huijun Wu who worked on lipopeptides of Bacillus and Pseudomonas at Nanjing Agricultural University.
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Natural Science Foundation of Jiangsu Province (BK 20201239) and the National Natural Science Foundation of China (NSFC 31201556, NSFC 32272624).
Author information
Authors and affiliations.
Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, Nanjing, 210014, Jiangsu, China
Junqing Qiao, Rongsheng Zhang, Youzhou Liu & Yongfeng Liu
Institute of Biology, Humboldt University Berlin, Berlin, Germany
Rainer Borriss
College of Plant Protection, Yangzhou University, Yangzhou, 225009, Jiangsu, China
Kai Sun & Xijun Chen
You can also search for this author in PubMed Google Scholar
Contributions
Junqing Qiao, Rainer Borriss, Youzhou Liu, Yongfeng Liu and Xijun Chen: conceived and summarized the content in the work; Kai Sun and Rongsheng Zhang performed the table and figures; Junqing Qiao, Rainer Borriss drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Correspondence to Rainer Borriss , Youzhou Liu or Yongfeng Liu .
Ethics declarations
Ethics approval and consent to participate, consent for publication, competing interests.
The authors declare that they have no competing interests.
Additional information
Publisher's note.
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ . The Creative Commons Public Domain Dedication waiver ( http://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
Reprints and permissions
About this article
Cite this article.
Qiao, J., Borriss, R., Sun, K. et al. Research advances in the identification of regulatory mechanisms of surfactin production by Bacillus : a review. Microb Cell Fact 23 , 100 (2024). https://doi.org/10.1186/s12934-024-02372-7
Download citation
Received : 16 November 2023
Accepted : 18 March 2024
Published : 02 April 2024
DOI : https://doi.org/10.1186/s12934-024-02372-7
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
- Regulatory mechanism
Microbial Cell Factories
ISSN: 1475-2859
- Submission enquiries: [email protected]

IMAGES
VIDEO
COMMENTS
3 10 Tips for Writing an Acknowledgement for a Research Paper. 4 5 Samples for Acknowledgment in Research Paper. 4.1 Sample 1: Acknowledgement for Collaborative Research: 4.2 Sample 2: Acknowledgement for Funding Support: 4.3 Sample 3: Acknowledgement for Mentorship and Guidance: 4.4 Sample 4: Acknowledgement for Institutional Support:
Formatting the acknowledgements. As a rule of thumb, the acknowledgement section should be a single short paragraph of say half a dozen lines. Examine the target journal for the format: whether the heading appears on a separate line or run on (that is, the text follows the heading on the same line). Check also whether the heading is in bold or ...
Acknowledgement in Research Paper - A Quick Guide [5 Examples] Academic. The acknowledgement section in your research paper is where you thank those who have helped or supported you throughout your research and writing. It is a short section of 3-5 paragraphs or no more than 300 words you put on a page after the title page.
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process. Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page. In your acknowledgements, it's okay to use a more ...
Writing the acknowledgements section of your thesis or dissertation is an opportunity to express gratitude to everyone who helped you along the way. Remember to: Acknowledge those people who significantly contributed to your research journey. Order your thanks from formal support to personal support. Maintain a balance between formal and ...
Begin your acknowledgements by expressing gratitude to those who have made the most significant contributions to your research. This could be your academic advisors, supervisors, or funding bodies. Starting with the most significant contributions helps to set the tone for the rest of your acknowledgements. Ensure that you express your gratitude ...
📋 Where do you put acknowledgements in a research paper? While acknowledgments are usually more present in academic theses, they can also be a part of research papers. In academic theses, acknowledgments are usually found at the beginning, somewhere between abstract and introduction. In research papers, acknowledgments are usually found at ...
Acknowledgments. This usually follows the Discussion and Conclusions sections. Its purpose is to thank all of the people who helped with the research but did not qualify for authorship (check the target journal's Instructions for Authors for authorship guidelines). Acknowledge anyone who provided intellectual assistance, technical help ...
Example 1. First and foremost I am extremely grateful to my supervisors, Prof. XXX and Dr. XXX for their invaluable advice, continuous support, and patience during my PhD study. Their immense knowledge and plentiful experience have encouraged me in all the time of my academic research and daily life.
help you write your Acknowledgements section of your dissertation. According to one source, the Acknowledgements section of a Ph.D. dissertation is the most widely read section. Whether you believe this or not, many individuals who helped you in the process of writing may check to see if, indeed, they have meant something to you.
The acknowledgment section helps identify the contributors responsible for specific parts of the project. It can include: Authors. Non-authors (colleagues, friends, supervisor, etc.) Funding sources. Editing services, Administrative staff. In academic writing, the information presented in the acknowledgment section should be kept brief.
This example of acknowledgements for a research paper is designed to demonstrate how intellectual, financial and other research contributions should be formally acknowledged in academic and scientific writing. As brief acknowledgements for a research paper, the example gathers contributions of different kinds - intellectual assistance ...
The acknowledgment section in a research paper credits individuals, institutions, or organizations that aided in the research or manuscript preparation. It's usually found after the conclusion. While optional, acknowledgments are commonly added to recognize and thank contributors for their efforts. This section typically starts with a ...
Finally, the ICMJE encourages written permission from acknowledged individuals "because acknowledgment may imply endorsement.". Funding sources should also be mentioned in the acknowledgments section, unless your target journal requires a separate section for this information. Whether the funding was partial or full, relevant grant numbers ...
Acknowledgement in a research paper is the section where the author expresses gratitude to individuals and organizations who have contributed to the completion of the study. This section is usually placed at the beginning or end of the paper and is an important part of the research process. It allows the author to recognize the support ...
Acknowledgment in a research paper is a section dedicated to expressing gratitude and recognizing the individuals, institutions, or resources that have contributed to the completion of the research. This section is an opportunity for the author to appreciate the support, guidance, or assistance received during the research process.
The acknowledgements section is your opportunity to thank those who have helped and supported you personally and professionally during your thesis or dissertation process. Thesis or dissertation acknowledgements appear between your title page and abstract and should be no longer than one page. In your acknowledgements, it's okay to use a more ...
The Acknowledgments section is present in both a paper and an academic thesis. For papers, the Acknowledgments section is usually presented at the back, whereas in a thesis, this section is located towards the front of the manuscript and is commonly placed somewhere between the abstract and Introduction.
In research papers, acknowledgements play a crucial role in recognizing the contributions and support of individuals or organizations who have helped in the completion of the study. They provide an opportunity to express gratitude towards people who provided guidance, resources, or encouragement during the research process. ...
Acknowledgements are usually a minor part of scientific papers, but they serve a very important function. This section of the manuscript is normally reserved to thank those who offered assistance, but not enough to merit authorship, funders, or any other people or organizations or artificial intelligent tools that may have in any way been directly associated either with the research reported ...
In fact, acknowledgement studies can no longer be separated from the financial aspect of scientific research. In 2008, WoS started to collect and index funding sources found in the acknowledgements of scientific papers. These new data were added by WoS in response to many funding bodies' requirement to acknowledge the sources supporting research.
Example 1: Acknowledgement. The completion of this research paper would not have been possible without the support and guidance of many individuals. I would like to express my deepest gratitude to my advisor, [Advisor Name], for their invaluable guidance, support, and encouragement throughout the entire research process.
Acknowledgements. Esin Durmus led the research, designed the experiments, ran the experiments, and analyzed the data. Esin Durmus and Liane Lovitt wrote the blog post. Jack Clark, Alex Tamkin, Liane Lovitt, Stuart Ritchie, and Deep Ganguli contributed to the experimental design and analysis, and gave feedback on the writing.
Acknowledgements. This research used data assets made available by ODAP as part of the Data and Connectivity National Core Study, led by Health Data Research UK in partnership with the Office for ...
In this paper I consider various shifts in my research and understanding stimulated by seeking how to combat social ecological crises connected to modern economies. The discussion and critical reflections are structured around five papers that were submitted to Environmental Values in an open call to address my work.
A multitude of regulators affect transcription of the srfAA-AD operon. The srfAA-AD operon transcribes the nonribosomal peptide synthetases and a thioesterase involved in surfactin synthesis (Fig. 1).The NRPS system consists of seven catalytic modules encoded by the genes srfAA, srfAB, srfAC, which is 27 kb in length [67, 68].Each catalytic module is responsible for the recognition and ...