Our website uses cookies, as almost all websites do, to help provide you with the best experience we can. Cookies are small text files that are placed on your computer or mobile phone when you browse websites. LEARN MORE >
Our website uses cookies, as almost all websites do, to help provide you with the best experience we can. Cookies are small text files that are placed on your computer or mobile phone when you browse websites.
Cookies help us:
- Make our website work as you’d expect.
- Provide a message we believe is more relevant to you.
We do not use cookies to:
- Collect any personally identifiable information.
- Collect any sensitive information.
- Pass personally identifiable data to third parties.
You can learn more about all the cookies and the information we collect by reading our Privacy Policy . If you don’t want to use cookies you can either exit the website or change your browser settings .
- What is All of Us

What All of Us can do for you
Why all of us needs you, how all of us protects your privacy.
The future of health begins with you
Too often, health care is one-size-fits-all. But imagine a future where prevention, treatment, and care are tailored for YOU.
That future starts with research that includes all of us.
What is All of Us?
Part of the National Institutes of Health , All of Us is changing how health research is done.
We're building one of the largest and most diverse health databases of its kind.
Researchers are already using this data to learn more about why people get sick or stay healthy, and what makes each of us unique.
They're using this information to find better ways to prevent and treat illnesses and to care for all of us.
But there's more to do.

- Your genetic ancestry
- Your risk for certain hereditary diseases
- Your body's reaction to certain medicines
- There is no cost to participate other than some of your time. Most people will spend no more than a few hours a year taking part in the program's activities.
- You can find out about research powered by the data you've shared.
- You will join a community of people who are already making a difference.
Many groups have been left out of health research in the past. You can help change that.
Researchers need information from large numbers of people who reflect the diversity of the United States. Our goal is to reach more than 1 million people from all backgrounds.
If you join All of Us , we will ask you to answer health surveys and connect your electronic health records and wearable devices. We may also ask you to share biosamples (like blood, urine, and saliva).
All of this information helps paint a full picture of what makes each of us unique. It helps researchers understand how our health history, genetics, environment, and life experiences impact our health.
Why join All of Us?
I joined the All of Us Research Program because I believe that health care should be as unique as each one of us and I want to be part of it.

Medical research has not always focused on minorities or Latinos. I think it's important for my generation to share our health information to help future generations.

I signed up for the All of Us Research Program because I represent a group that has historically been underrepresented in research and I want to be counted.

I believe firmly that the All of Us Research Program will be the first step in generating new discoveries that will translate into patient care.

How to Participate
You are more than welcome to only sign the consent to join. You will still be able to participate by answering surveys and taking part in other activities.
However, you won’t be invited to provide your sample (blood or saliva) or you won’t get the $25 compensation for your time. We won’t be able to offer you your DNA results.
If you have an in-person appointment, you will have your measurements taken:
- Hip circumference.
- Waist circumference.
- Blood pressure.
- Heart rate.
You will also be able to provide a sample (blood or saliva). You can still provide a sample without agreeing to receive your DNA results.
You will receive $25 if you agree to share your Electronic Health Records (EHRs) and have your in-person visit to provide a sample (blood or saliva) and/or measurements.
- Your genetic ancestry.
- Your hereditary disease risk.
- How your body may react to certain medicines.
You’ll get your DNA results if you complete a series of activities:
- Agree to share your Electronic Health Records (EHRs).
- Say yes to get your DNA results.
- Finish “The Basics” survey.
- Provide a sample (blood or saliva).
- Verify your identity.
It might take a few months or years to receive your DNA results. In some cases, we might not be able to offer them to you because:
- We were not able to get enough DNA from your sample to study it.
- You had a bone marrow transplant.
- We cannot verify your identity.
- Answer more surveys.
- Connect a wearable device (e.g., Fitbit, Apple Watch).
- Learn about other research opportunities.
All of Us is a research program and does not provide health care or medical advice.
All of Us follows all laws and rules for keeping the data you share with us safe and private.
We often test the security of our databases. Before making data available to researchers, we remove personal details that could identify you. We also require researchers to go through training. They must agree to our data use rules.
Voices of All of Us

All of Us requires training to make sure that every researcher who wants to use the Workbench understands how to really follow the rules set in place by All of Us .

The safety, the security, and the ethics of this program are at the highest level. We really care about our participants as partners.

It’s an easy thing to do, it’s safe, it’s secure, and I hope you’ll be involved too.

They do a great job of protecting people who may disclose things through the All of Us data set ... that may not necessarily be disclosed in their day-to-day lives.
Learn more from the Voices of All of Us

Where All of Us is now
All of Us celebrates its 5-year anniversary in May 2023 .
So far, more than 780,000 people have joined the program. More than 80% of All of Us participants are from groups that have rarely been part of health research.
Help us reach 1,000,000+ participants
*Fully enrolled participants are those who have shared their health information with All of Us , including giving blood and urine or saliva samples.
Last updated: March 27, 2024
Are you ready to help shape the future of health?
All of Us works closely with Health Care Provider Organizations around the country to make joining easy.
People who live too far from a partner site can join online.
Sign up online
Sign up in-person
Join an All of Us Event
If you prefer, you can call (844) 842-2855 and a support center guide can walk you through the sign up process over the phone.
We’re open from 7 a.m. to 10 p.m. Eastern, excluding public holidays. Toll-free TTY-based Telecommunications Relay Service is available by dialing 711.
Your participation in All of Us is voluntary. You can choose to leave the program at any time.
- (844) 842-2855
- TTY dial 711
- [email protected]

What is the All of Us Research Program?
All of Us is a Research Program of the National Institutes of Health (NIH). The mission of All of Us is to accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us by building dataset of one million or more volunteers nationwide who will sign up to share their health information over time.
All of Us Enrollment and Engagement Partners
To reach its goal, the All of Us Research Program is partnering with leading institutions, organizations, community partners, and participant representatives across the country.
Bringing together public libraries & All of Us
The NAPC provides funding to public libraries to build and strengthen partnerships with All of Us Enrollment and Engagement Partners through a variety of activities.
How can public libraries support All of Us ?
- Share informational materials about All of Us on your community boards.
- Offer space at your library for All of Us tabling outreach.
- Invite All of Us to participate at library programs like health fairs, community festivals, and open house events.
- Host an expert from All of Us for a program about a health topic important to your community.
- Propose your own project!
Interested working with your local All of Us Enrollment and Engagement Partners?
Email the NAPC at [email protected] .

Our website uses cookies, as almost all websites do, to help provide you with the best experience we can. Cookies are small text files that are placed on your computer or mobile phone when you browse websites. LEARN MORE >
Cookies help us:
- Make our website work as you’d expect.
- Provide a message we believe is more relevant to you.
We do not use cookies to:
- Collect any personally identifiable information.
- Collect any sensitive information.
- Pass personally identifiable data to third parties.
You can learn more about all the cookies and the information we collect by reading our Privacy Policy . If you don’t want to use cookies you can either exit the website or change your browser settings .
- How It Works
Learn about the All of Us WEAR Study - if eligible, you could get a Fitbit at no cost!
Health discoveries come from research. Research starts with you .
Join the largest and most inclusive health research initiative of its kind. You could help researchers find answers to the most pressing health questions.

Join over 700,000 people and counting.
All of Us , a research program from the National Institutes of Health (NIH), aims to have one million or more volunteers who reflect the diversity of the United States. Researchers will use the participant data collected to study many different conditions, including cancer, diabetes, depression, asthma, and other diseases.

Personally, I wanted to become a participant in this program and be involved because, as a black person living in America, I know that we’ve been left out of biohealth research largely in the past.
– Shantelle , All of Us participant
I represent a group that has historically been unrepresented in research, and I want to be counted.
– Keisha , All of Us participant
It is very important for my generation to share their health information to help future generations and help the population as a whole.
– Carlos , All of Us participant

Sometimes the smallest actions can have the largest impact.
Without you, it won’t be All of Us . You could help improve the lives of future generations. For your family. For your community. For our future.

Research starts with you.
All of Us has an ambitious goal: We want to help researchers discover future medical breakthroughs. To do that, we’re building a modern data hub. This will power health research from doctors and researchers all over the world.
Right now, health data is hard to access, hard to search, and is not diverse enough to represent the U.S. population. All of Us wants to change that by creating the largest health research database of its kind. This will help researchers find and access the information they need. Then, they can spend more time on what’s really important—using research to find answers and make discoveries.
Diversity matters in research.
Hispanics make up 18.5% of the U.S. population.
Yet only 3% of Hispanics are represented in industry- sponsored studies.
We're CHANGING that.
17.5% of All of Us participants are Hispanic.
There is more left to accomplish. All of Us is focused on giving health researchers the best information possible to make new discoveries. Help drive research for a healthier future.

Ready to make a difference?
Meet our partners.
The All of Us Research Program partners with leading institutions, organizations, community partners, and participant representatives across the country. Our partnerships include:

The All of Us Research Program WEAR Study

The All of Us Research Program’s WEAR Study has begun! If eligible, you could be part of the WEAR Study and receive a new Fitbit wearable device at no cost to you. All of Us will receive the data that Fitbit collects. This data may help us understand how behavior impacts health.
Download the All of Us App
Looking for more.
Sign up and we’ll send you more information about our groundbreaking research program!
Thank you for subscribing!
By clicking, you agree to the terms described in our Privacy Policy & Terms of Service. Message & data rates may apply.
Our advisors are ready to answer your questions. We’re open from 7 a.m. to 10 p.m. Eastern, excluding public holidays.
Toll-free TTY-based Telecommunications Relay Service is available by dialing 711.
844-842-2855
[email protected]
Your privacy is important to us. For security, please do not include any personal information (address, social security number, or health details) in your email or chat messages.
The All of Us Research Program is on a mission to help change health research to represent our country’s diversity. We’re collecting health data from over a million people in an effort to help researchers learn more about health.
All of Us and the All of Us logo are service marks of the U.S. Department of Health Services. The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
Copyright ©2023
Privacy policy & terms
All of Us: Release of Nearly 100,000 Whole Genome Sequences Sets Stage for New Discoveries
Posted on March 29th, 2022 by Joshua Denny, M.D., M.S., and Lawrence Tabak, D.D.S., Ph.D.

Nearly four years ago, NIH opened national enrollment for the All of Us Research Program . This historic program is building a vital research community within the United States of at least 1 million participant partners from all backgrounds. Its unifying goal is to advance precision medicine, an emerging form of health care tailored specifically to the individual, not the average patient as is now often the case. As part of this historic effort, many participants have offered DNA samples for whole genome sequencing, which provides information about almost all of an individual’s genetic makeup.
Earlier this month, the All of Us Research Program hit an important milestone. We released the first set of nearly 100,000 whole genome sequences from our participant partners. The sequences are stored in the All of Us Researcher Workbench , a powerful, cloud-based analytics platform that makes these data broadly accessible to registered researchers.
The All of Us Research Program and its many participant partners are leading the way toward more equitable representation in medical research. About half of this new genomic information comes from people who self-identify with a racial or ethnic minority group. That’s extremely important because, until now, over 90 percent of participants in large genomic studies were of European descent. This lack of diversity has had huge impacts—deepening health disparities and hindering scientific discovery from fully benefiting everyone.
The Researcher Workbench also contains information from many of the participants’ electronic health records, Fitbit devices, and survey responses. Another neat feature is that the platform links to data from the U.S. Census Bureau’s American Community Survey to provide more details about the communities where participants live.
This unique and comprehensive combination of data will be key in transforming our understanding of health and disease. For example, given the vast amount of data and diversity in the Researcher Workbench, new diseases are undoubtedly waiting to be uncovered and defined . Many new genetic variants are also waiting to be identified that may better predict disease risk and response to treatment .
To speed up the discovery process, these data are being made available, both widely and wisely. To protect participants’ privacy, the program has removed all direct identifiers from the data and upholds strict requirements for researchers seeking access. Already, more than 1,500 scientists across the United States have gained access to the Researcher Workbench through their institutions after completing training and agreeing to the program’s strict rules for responsible use. Some of these researchers are already making discoveries that promote precision medicine, such as finding ways to predict how to best to prevent vision loss in patients with glaucoma .
Beyond making genomic data available for research, All of Us participants have the opportunity to receive their personal DNA results , at no cost to them. So far, the program has offered genetic ancestry and trait results to more than 100,000 participants. Plans are underway to begin sharing health-related DNA results on hereditary disease risk and medication-gene interactions later this year.
This first release of genomic data is a huge milestone for the program and for health research more broadly, but it’s also just the start. The program’s genome centers continue to generate the genomic data and process about 5,000 additional participant DNA samples every week.
The ultimate goal is to gather health data from at least 1 million or more people living in the United States, and there’s plenty of time to join the effort. Whether you would like to contribute your own DNA and health information, engage in research, or support the All of Us Research Program as a partner, it’s easy to get involved. By taking part in this historic program, you can help to build a better and more equitable future for health research and precision medicine.
Note: Joshua Denny, M.D., M.S. , is the Chief Executive Officer of NIH’s All of Us Research Program.
All of Us Research Program (NIH)
All of Us Research Hub
Join All of Us (NIH)
Share this:
- Click to share on Facebook (Opens in new window)
- Click to share on Twitter (Opens in new window)
- Click to share on LinkedIn (Opens in new window)
- Click to share on Pinterest (Opens in new window)
- Click to share on Tumblr (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to share on Telegram (Opens in new window)
- Click to share on WhatsApp (Opens in new window)
- Click to print (Opens in new window)
Posted In: News
Tags: All of Us , All of Us Research Program , All of Us Researcher Workbench , big data , cohort , data protection , diversity , DNA , DNA sequencing , EHR , electronic health records , ethnicity , Fitbit , genome , genomics , glaucoma , health disparities , precision medicine , race , research privacy , U.S. Census Bureau , whole genome sequencing
Wow! What an accomplishment
I hope there are a diverse group of ethics people involved as well. Information misused for eugenics type projects have dire consequences. Given the ubiquitous nature of food allergies and CYP450 metabolism, not everyone on this planet will use the information for “altruistic” purposes. Trust is all well and good until it is abused.
This is very useful
Wow! What an accomplishment …
Leave a Comment Cancel reply
@nihdirector on twitter, nih on facebook.
Kendall Morgan, Ph.D.
Comments and Questions
If you have comments or questions not related to the current discussions, please direct them to Ask NIH .
You are encouraged to share your thoughts and ideas. Please review the NIH Comments Policy

- Visitor Information
- Privacy Notice
- Accessibility
- No Fear Act
- HHS Vulnerability Disclosure
- U.S. Department of Health and Human Services
- USA.gov – Government Made Easy
Discover more from NIH Director's Blog
Subscribe now to keep reading and get access to the full archive.
Type your email…
Continue reading

About the All of Us Research Program
The National Institutes of Health’s All of Us Research Program is a historic effort to collect and study data from at least one million people living in the United States. The goal of All of Us is to speed up health research discoveries, enabling new kinds of individualized health care. To make this possible, the program is building one of the world’s largest and most diverse databases for health research. The program aims to reflect the diversity of the United States and to include participants from communities that have been underrepresented in health research in the past.
The All of Us Research Program is dedicated to providing support to institutions that have a documented historical mission or historical commitment to training underrepresented students. The program recognizes the important role these institutions have played in supporting scientific research, particularly on diseases or conditions that disproportionately impact racial and ethnic minorities and other U.S. populations that experience health disparities. Although these institutions are uniquely positioned to engage underrepresented populations in research and in the translation of research advances into culturally competent, measurable, and sustained improvements in health outcomes, they often lack sufficient capacity to conduct and sustain cutting-edge health-related research.
Subscribe to the Academy's Connections Newsletter
Read our quarterly newsletter to receive news about academy researchers, opportunities, events, and course announcements
Read past issues here
About the All of Us Researcher Academy
The All of Us Researcher Academy is a comprehensive program that provides training and technical assistance for researchers who are conducting research with the All of Us Researcher Workbench —the cloud-based analytics platform where registered researchers can access data from All of Us participants . The academy also supports peer-to-peer learning and network-building among researchers and students.
The academy is dedicated to providing support to institutions that have a documented historical mission or historical commitment to training underrepresented students. Currently, the All of Us Researcher Academy resources are free for students, faculty, and post-docs at institutions with a track record of training researchers underrepresented in the biomedical workforce. See the Notice of NIH’s Interest in Diversity for examples of groups that have been shown to be underrepresented in the biomedical research workforce.
RTI International leads the All of Us Researcher Academy in collaboration with the All of Us Research Program’s Division of Engagement and Outreach. The division partners with community organizations nationwide to foster relationships with participants, researchers, and health care providers. The academy and other researcher engagement activities are central to building a diverse community of researchers.
Read our quarterly newsletter to receive news about academy researchers, opportunities, events, and course announcements.
Click here to read past issues.
Get Involved with the Researcher Academy
- On September 6, Community-Campus Partnerships for Health will host a Researcher Academy Road Tour event at the Morehouse School of Medicine in Atlanta, GA. The event, “Utilizing the All of Us Researcher Workbench at HBCUs for Community Health Research,” is from 10:00 am–12:00 pm ET. Register here to attend in person or remotely.
Mentor and Internship Opportunities
Apply online to become an All of Us Researcher Academy intern or mentor:
- Applications for the Researcher Academy Internship Program are now closed.
- Intern application
- Mentor application
Institutional Opportunities
- Applications for Institutional Champion Awards are now closed.
- Applications for Institutional Champion Awards are now open. Access the Call for Applications and link to the electronic application form here . Applications are due by 12:00 pm ET, January 5, 2024.
- Applications for the Institutional Champion award are closed.
- Hispanic-Serving Institutions can learn more and complete a Request for Application to receive funding as a Researcher Academy Institutional Champion by noon on September 15, 2023.

Course Offerings
The academy resources are available at no cost to all registered researchers based on availability. Registration is open for the following courses:
Introduction to Publishing Health Data in Academic Journals
Live virtual course April 4, 2024, 3:00–4:15 pm ET and April 11, April 18, May 2, 2024, 3:00–4:00 pm ET
Instructor: Dr. Vabren Watts
A short course series helping to guide participants in the academy with their career paths and informing them of requirements and opportunities that are essential for their success in STEM or STEM-related research. This course is ideal for undergraduate or graduate students interested in publishing their research for the first time.
Register for this course
Mindfulness Practices for Health Researchers
Live virtual course April 22–25, 2024; 5:15–5:45 pm ET
Instructor: Dr. TJ Exford
This course will lead participants through a progression of mindfulness exercises for health researchers. Participants will learn to intentionally focus their attention, understand what mindfulness is and how it makes a difference, and be able to use mindful attitudes like acceptance, kindness, gratitude, and loving-kindness. This course is ideal for everyone.
Past Webinars
- All of Us Researcher Academy HBCU Road Tour at Tuskegee University (April 2023)
- All of Us Researcher Academy Celebration of the First Cohort of Institutional Champion Awardees/HBCU Road Tour at Howard University (January 2023)
- Introducing the All of Us Researcher Academy (June 2022)
Press Releases
- All of Us Researcher Academy Names Third Group of Institutional Champions
- All of Us Researcher Academy Selects Second Cohort of Institutional Champions
- Introducing the NIH-Supported All of Us Researcher Academy
- All of Us Researcher Academy Selects 6 HBCUs as Institutional Champions

Questions about the All of Us Researcher Academy?
Contact [email protected] to learn how you can participate.
This platform is developed by RTI International . The All of Us Researcher Academy is supported by the Division of Engagement and Outreach, All of Us Research Program, National Institutes of Health, Award Number 1OT20D028395-01.
All of Us and the All of Us logo are registered service marks of the U.S. Department of Health and Human Services . The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
Explore the data at ResearchAllofUs.org
Learn more about the program protocol, leadership, and governance at AllofUs.nih.gov
Learn more about becoming a participant at JoinAllofUs.org
Copyright © 2022 Privacy Policy Terms -->
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Open access
- Published: 19 February 2024
Genomic data in the All of Us Research Program
The all of us research program genomics investigators.
Nature volume 627 , pages 340–346 ( 2024 ) Cite this article
110k Accesses
3 Citations
1096 Altmetric
Metrics details
- Genetic variation
Genome-wide association studies
Comprehensively mapping the genetic basis of human disease across diverse individuals is a long-standing goal for the field of human genetics 1 , 2 , 3 , 4 . The All of Us Research Program is a longitudinal cohort study aiming to enrol a diverse group of at least one million individuals across the USA to accelerate biomedical research and improve human health 5 , 6 . Here we describe the programme’s genomics data release of 245,388 clinical-grade genome sequences. This resource is unique in its diversity as 77% of participants are from communities that are historically under-represented in biomedical research and 46% are individuals from under-represented racial and ethnic minorities. All of Us identified more than 1 billion genetic variants, including more than 275 million previously unreported genetic variants, more than 3.9 million of which had coding consequences. Leveraging linkage between genomic data and the longitudinal electronic health record, we evaluated 3,724 genetic variants associated with 117 diseases and found high replication rates across both participants of European ancestry and participants of African ancestry. Summary-level data are publicly available, and individual-level data can be accessed by researchers through the All of Us Researcher Workbench using a unique data passport model with a median time from initial researcher registration to data access of 29 hours. We anticipate that this diverse dataset will advance the promise of genomic medicine for all.
Similar content being viewed by others

Inferring gene regulatory networks from single-cell multiome data using atlas-scale external data
Qiuyue Yuan & Zhana Duren

Emil Uffelmann, Qin Qin Huang, … Danielle Posthuma

Tissue-specific enhancer–gene maps from multimodal single-cell data identify causal disease alleles
Saori Sakaue, Kathryn Weinand, … Soumya Raychaudhuri
Comprehensively identifying genetic variation and cataloguing its contribution to health and disease, in conjunction with environmental and lifestyle factors, is a central goal of human health research 1 , 2 . A key limitation in efforts to build this catalogue has been the historic under-representation of large subsets of individuals in biomedical research including individuals from diverse ancestries, individuals with disabilities and individuals from disadvantaged backgrounds 3 , 4 . The All of Us Research Program (All of Us) aims to address this gap by enrolling and collecting comprehensive health data on at least one million individuals who reflect the diversity across the USA 5 , 6 . An essential component of All of Us is the generation of whole-genome sequence (WGS) and genotyping data on one million participants. All of Us is committed to making this dataset broadly useful—not only by democratizing access to this dataset across the scientific community but also to return value to the participants themselves by returning individual DNA results, such as genetic ancestry, hereditary disease risk and pharmacogenetics according to clinical standards, to those who wish to receive these research results.
Here we describe the release of WGS data from 245,388 All of Us participants and demonstrate the impact of this high-quality data in genetic and health studies. We carried out a series of data harmonization and quality control (QC) procedures and conducted analyses characterizing the properties of the dataset including genetic ancestry and relatedness. We validated the data by replicating well-established genotype–phenotype associations including low-density lipoprotein cholesterol (LDL-C) and 117 additional diseases. These data are available through the All of Us Researcher Workbench, a cloud platform that embodies and enables programme priorities, facilitating equitable data and compute access while ensuring responsible conduct of research and protecting participant privacy through a passport data access model.
The All of Us Research Program
To accelerate health research, All of Us is committed to curating and releasing research data early and often 6 . Less than five years after national enrolment began in 2018, this fifth data release includes data from more than 413,000 All of Us participants. Summary data are made available through a public Data Browser, and individual-level participant data are made available to researchers through the Researcher Workbench (Fig. 1a and Data availability).
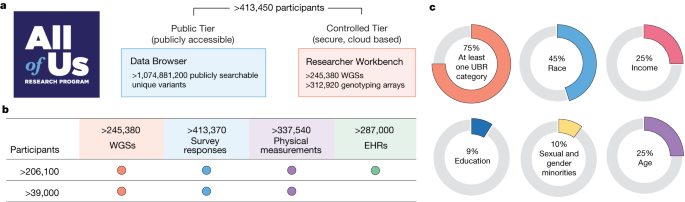
a , The All of Us Research Hub contains a publicly accessible Data Browser for exploration of summary phenotypic and genomic data. The Researcher Workbench is a secure cloud-based environment of participant-level data in a Controlled Tier that is widely accessible to researchers. b , All of Us participants have rich phenotype data from a combination of physical measurements, survey responses, EHRs, wearables and genomic data. Dots indicate the presence of the specific data type for the given number of participants. c , Overall summary of participants under-represented in biomedical research (UBR) with data available in the Controlled Tier. The All of Us logo in a is reproduced with permission of the National Institutes of Health’s All of Us Research Program.
Participant data include a rich combination of phenotypic and genomic data (Fig. 1b ). Participants are asked to complete consent for research use of data, sharing of electronic health records (EHRs), donation of biospecimens (blood or saliva, and urine), in-person provision of physical measurements (height, weight and blood pressure) and surveys initially covering demographics, lifestyle and overall health 7 . Participants are also consented for recontact. EHR data, harmonized using the Observational Medical Outcomes Partnership Common Data Model 8 ( Methods ), are available for more than 287,000 participants (69.42%) from more than 50 health care provider organizations. The EHR dataset is longitudinal, with a quarter of participants having 10 years of EHR data (Extended Data Fig. 1 ). Data include 245,388 WGSs and genome-wide genotyping on 312,925 participants. Sequenced and genotyped individuals in this data release were not prioritized on the basis of any clinical or phenotypic feature. Notably, 99% of participants with WGS data also have survey data and physical measurements, and 84% also have EHR data. In this data release, 77% of individuals with genomic data identify with groups historically under-represented in biomedical research, including 46% who self-identify with a racial or ethnic minority group (Fig. 1c , Supplementary Table 1 and Supplementary Note ).
Scaling the All of Us infrastructure
The genomic dataset generated from All of Us participants is a resource for research and discovery and serves as the basis for return of individual health-related DNA results to participants. Consequently, the US Food and Drug Administration determined that All of Us met the criteria for a significant risk device study. As such, the entire All of Us genomics effort from sample acquisition to sequencing meets clinical laboratory standards 9 .
All of Us participants were recruited through a national network of partners, starting in 2018, as previously described 5 . Participants may enrol through All of Us - funded health care provider organizations or direct volunteer pathways and all biospecimens, including blood and saliva, are sent to the central All of Us Biobank for processing and storage. Genomics data for this release were generated from blood-derived DNA. The programme began return of actionable genomic results in December 2022. As of April 2023, approximately 51,000 individuals were sent notifications asking whether they wanted to view their results, and approximately half have accepted. Return continues on an ongoing basis.
The All of Us Data and Research Center maintains all participant information and biospecimen ID linkage to ensure that participant confidentiality and coded identifiers (participant and aliquot level) are used to track each sample through the All of Us genomics workflow. This workflow facilitates weekly automated aliquot and plating requests to the Biobank, supplies relevant metadata for the sample shipments to the Genome Centers, and contains a feedback loop to inform action on samples that fail QC at any stage. Further, the consent status of each participant is checked before sample shipment to confirm that they are still active. Although all participants with genomic data are consented for the same general research use category, the programme accommodates different preferences for the return of genomic data to participants and only data for those individuals who have consented for return of individual health-related DNA results are distributed to the All of Us Clinical Validation Labs for further evaluation and health-related clinical reporting. All participants in All of Us that choose to get health-related DNA results have the option to schedule a genetic counselling appointment to discuss their results. Individuals with positive findings who choose to obtain results are required to schedule an appointment with a genetic counsellor to receive those findings.
Genome sequencing
To satisfy the requirements for clinical accuracy, precision and consistency across DNA sample extraction and sequencing, the All of Us Genome Centers and Biobank harmonized laboratory protocols, established standard QC methodologies and metrics, and conducted a series of validation experiments using previously characterized clinical samples and commercially available reference standards 9 . Briefly, PCR-free barcoded WGS libraries were constructed with the Illumina Kapa HyperPrep kit. Libraries were pooled and sequenced on the Illumina NovaSeq 6000 instrument. After demultiplexing, initial QC analysis is performed with the Illumina DRAGEN pipeline (Supplementary Table 2 ) leveraging lane, library, flow cell, barcode and sample level metrics as well as assessing contamination, mapping quality and concordance to genotyping array data independently processed from a different aliquot of DNA. The Genome Centers use these metrics to determine whether each sample meets programme specifications and then submits sequencing data to the Data and Research Center for further QC, joint calling and distribution to the research community ( Methods ).
This effort to harmonize sequencing methods, multi-level QC and use of identical data processing protocols mitigated the variability in sequencing location and protocols that often leads to batch effects in large genomic datasets 9 . As a result, the data are not only of clinical-grade quality, but also consistent in coverage (≥30× mean) and uniformity across Genome Centers (Supplementary Figs. 1 – 5 ).
Joint calling and variant discovery
We carried out joint calling across the entire All of Us WGS dataset (Extended Data Fig. 2 ). Joint calling leverages information across samples to prune artefact variants, which increases sensitivity, and enables flagging samples with potential issues that were missed during single-sample QC 10 (Supplementary Table 3 ). Scaling conventional approaches to whole-genome joint calling beyond 50,000 individuals is a notable computational challenge 11 , 12 . To address this, we developed a new cloud variant storage solution, the Genomic Variant Store (GVS), which is based on a schema designed for querying and rendering variants in which the variants are stored in GVS and rendered to an analysable variant file, as opposed to the variant file being the primary storage mechanism (Code availability). We carried out QC on the joint call set on the basis of the approach developed for gnomAD 3.1 (ref. 13 ). This included flagging samples with outlying values in eight metrics (Supplementary Table 4 , Supplementary Fig. 2 and Methods ).
To calculate the sensitivity and precision of the joint call dataset, we included four well-characterized samples. We sequenced the National Institute of Standards and Technology reference materials (DNA samples) from the Genome in a Bottle consortium 13 and carried out variant calling as described above. We used the corresponding published set of variant calls for each sample as the ground truth in our sensitivity and precision calculations 14 . The overall sensitivity for single-nucleotide variants was over 98.7% and precision was more than 99.9%. For short insertions or deletions, the sensitivity was over 97% and precision was more than 99.6% (Supplementary Table 5 and Methods ).
The joint call set included more than 1 billion genetic variants. We annotated the joint call dataset on the basis of functional annotation (for example, gene symbol and protein change) using Illumina Nirvana 15 . We defined coding variants as those inducing an amino acid change on a canonical ENSEMBL transcript and found 272,051,104 non-coding and 3,913,722 coding variants that have not been described previously in dbSNP 16 v153 (Extended Data Table 1 ). A total of 3,912,832 (99.98%) of the coding variants are rare (allelic frequency < 0.01) and the remaining 883 (0.02%) are common (allelic frequency > 0.01). Of the coding variants, 454 (0.01%) are common in one or more of the non-European computed ancestries in All of Us, rare among participants of European ancestry, and have an allelic number greater than 1,000 (Extended Data Table 2 and Extended Data Fig. 3 ). The distributions of pathogenic, or likely pathogenic, ClinVar variant counts per participant, stratified by computed ancestry, filtered to only those variants that are found in individuals with an allele count of <40 are shown in Extended Data Fig. 4 . The potential medical implications of these known and new variants with respect to variant pathogenicity by ancestry are highlighted in a companion paper 17 . In particular, we find that the European ancestry subset has the highest rate of pathogenic variation (2.1%), which was twice the rate of pathogenic variation in individuals of East Asian ancestry 17 .The lower frequency of variants in East Asian individuals may be partially explained by the fact the sample size in that group is small and there may be knowledge bias in the variant databases that is reducing the number of findings in some of the less-studied ancestry groups.
Genetic ancestry and relatedness
Genetic ancestry inference confirmed that 51.1% of the All of Us WGS dataset is derived from individuals of non-European ancestry. Briefly, the ancestry categories are based on the same labels used in gnomAD 18 . We trained a classifier on a 16-dimensional principal component analysis (PCA) space of a diverse reference based on 3,202 samples and 151,159 autosomal single-nucleotide polymorphisms. We projected the All of Us samples into the PCA space of the training data, based on the same single-nucleotide polymorphisms from the WGS data, and generated categorical ancestry predictions from the trained classifier ( Methods ). Continuous genetic ancestry fractions for All of Us samples were inferred using the same PCA data, and participants’ patterns of ancestry and admixture were compared to their self-identified race and ethnicity (Fig. 2 and Methods ). Continuous ancestry inference carried out using genome-wide genotypes yields highly concordant estimates.
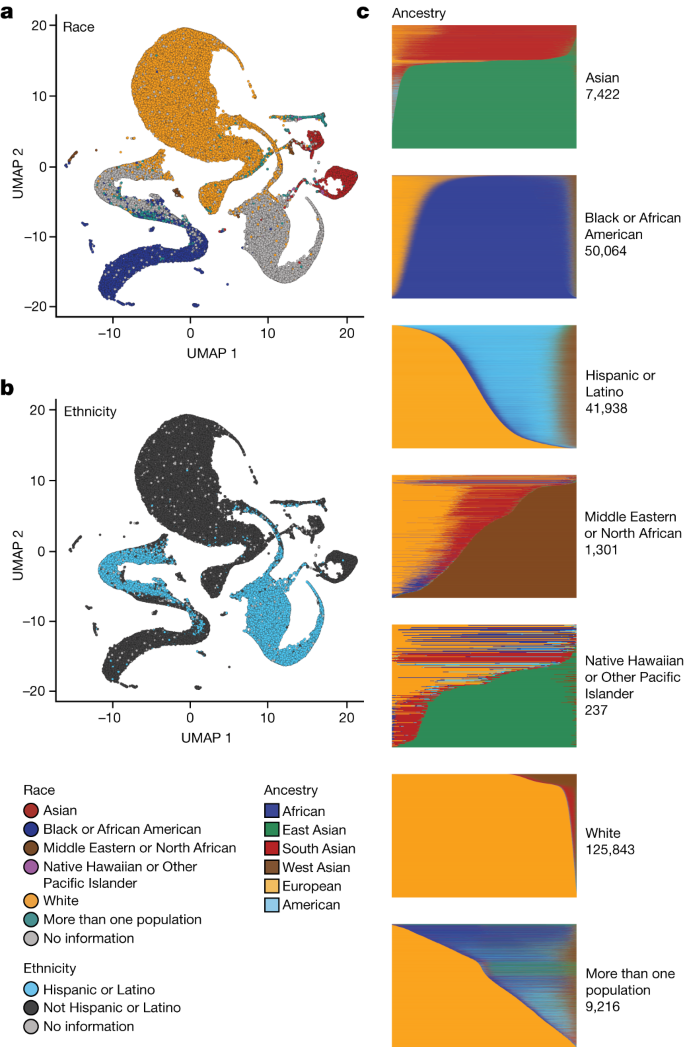
a , b , Uniform manifold approximation and projection (UMAP) representations of All of Us WGS PCA data with self-described race ( a ) and ethnicity ( b ) labels. c , Proportion of genetic ancestry per individual in six distinct and coherent ancestry groups defined by Human Genome Diversity Project and 1000 Genomes samples.
Kinship estimation confirmed that All of Us WGS data consist largely of unrelated individuals with about 85% (215,107) having no first- or second-degree relatives in the dataset (Supplementary Fig. 6 ). As many genomic analyses leverage unrelated individuals, we identified the smallest set of samples that are required to be removed from the remaining individuals that had first- or second-degree relatives and retained one individual from each kindred. This procedure yielded a maximal independent set of 231,442 individuals (about 94%) with genome sequence data in the current release ( Methods ).
Genetic determinants of LDL-C
As a measure of data quality and utility, we carried out a single-variant genome-wide association study (GWAS) for LDL-C, a trait with well-established genomic architecture ( Methods ). Of the 245,388 WGS participants, 91,749 had one or more LDL-C measurements. The All of Us LDL-C GWAS identified 20 well-established genome-wide significant loci, with minimal genomic inflation (Fig. 3 , Extended Data Table 3 and Supplementary Fig. 7 ). We compared the results to those of a recent multi-ethnic LDL-C GWAS in the National Heart, Lung, and Blood Institute (NHLBI) TOPMed study that included 66,329 ancestrally diverse (56% non-European ancestry) individuals 19 . We found a strong correlation between the effect estimates for NHLBI TOPMed genome-wide significant loci and those of All of Us ( R 2 = 0.98, P < 1.61 × 10 −45 ; Fig. 3 , inset). Notably, the per-locus effect sizes observed in All of Us are decreased compared to those in TOPMed, which is in part due to differences in the underlying statistical model, differences in the ancestral composition of these datasets and differences in laboratory value ascertainment between EHR-derived data and epidemiology studies. A companion manuscript extended this work to identify common and rare genetic associations for three diseases (atrial fibrillation, coronary artery disease and type 2 diabetes) and two quantitative traits (height and LDL-C) in the All of Us dataset and identified very high concordance with previous efforts across all of these diseases and traits 20 .
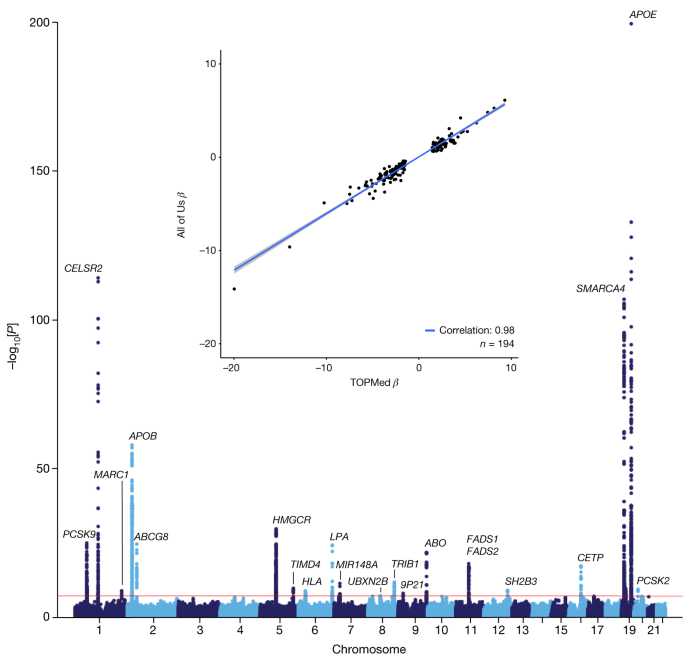
Manhattan plot demonstrating robust replication of 20 well-established LDL-C genetic loci among 91,749 individuals with 1 or more LDL-C measurements. The red horizontal line denotes the genome wide significance threshold of P = 5 × 10 –8 . Inset, effect estimate ( β ) comparison between NHLBI TOPMed LDL-C GWAS ( x axis) and All of Us LDL-C GWAS ( y axis) for the subset of 194 independent variants clumped (window 250 kb, r2 0.5) that reached genome-wide significance in NHLBI TOPMed.
Genotype-by-phenotype associations
As another measure of data quality and utility, we tested replication rates of previously reported phenotype–genotype associations in the five predicted genetic ancestry populations present in the Phenotype/Genotype Reference Map (PGRM): AFR, African ancestry; AMR, Latino/admixed American ancestry; EAS, East Asian ancestry; EUR, European ancestry; SAS, South Asian ancestry. The PGRM contains published associations in the GWAS catalogue in these ancestry populations that map to International Classification of Diseases-based phenotype codes 21 . This replication study specifically looked across 4,947 variants, calculating replication rates for powered associations in each ancestry population. The overall replication rates for associations powered at 80% were: 72.0% (18/25) in AFR, 100% (13/13) in AMR, 46.6% (7/15) in EAS, 74.9% (1,064/1,421) in EUR, and 100% (1/1) in SAS. With the exception of the EAS ancestry results, these powered replication rates are comparable to those of the published PGRM analysis where the replication rates of several single-site EHR-linked biobanks ranges from 76% to 85%. These results demonstrate the utility of the data and also highlight opportunities for further work understanding the specifics of the All of Us population and the potential contribution of gene–environment interactions to genotype–phenotype mapping and motivates the development of methods for multi-site EHR phenotype data extraction, harmonization and genetic association studies.
More broadly, the All of Us resource highlights the opportunities to identify genotype–phenotype associations that differ across diverse populations 22 . For example, the Duffy blood group locus ( ACKR1 ) is more prevalent in individuals of AFR ancestry and individuals of AMR ancestry than in individuals of EUR ancestry. Although the phenome-wide association study of this locus highlights the well-established association of the Duffy blood group with lower white blood cell counts both in individuals of AFR and AMR ancestry 23 , 24 , it also revealed genetic-ancestry-specific phenotype patterns, with minimal phenotypic associations in individuals of EAS ancestry and individuals of EUR ancestry (Fig. 4 and Extended Data Table 4 ). Conversely, rs9273363 in the HLA-DQB1 locus is associated with increased risk of type 1 diabetes 25 , 26 and diabetic complications across ancestries, but only associates with increased risk of coeliac disease in individuals of EUR ancestry (Extended Data Fig. 5 ). Similarly, the TCF7L2 locus 27 strongly associates with increased risk of type 2 diabetes and associated complications across several ancestries (Extended Data Fig. 6 ). Association testing results are available in Supplementary Dataset 1 .
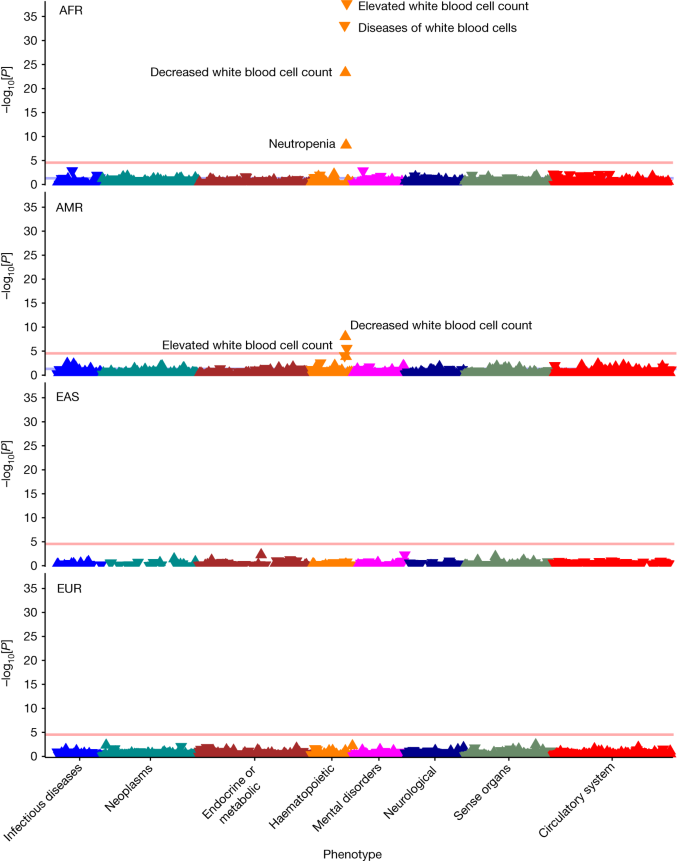
Results of genetic-ancestry-stratified phenome-wide association analysis among unrelated individuals highlighting ancestry-specific disease associations across the four most common genetic ancestries of participant. Bonferroni-adjusted phenome-wide significance threshold (<2.88 × 10 −5 ) is plotted as a red horizontal line. AFR ( n = 34,037, minor allele fraction (MAF) 0.82); AMR ( n = 28,901, MAF 0.10); EAS ( n = 32,55, MAF 0.003); EUR ( n = 101,613, MAF 0.007).
The cloud-based Researcher Workbench
All of Us genomic data are available in a secure, access-controlled cloud-based analysis environment: the All of Us Researcher Workbench. Unlike traditional data access models that require per-project approval, access in the Researcher Workbench is governed by a data passport model based on a researcher’s authenticated identity, institutional affiliation, and completion of self-service training and compliance attestation 28 . After gaining access, a researcher may create a new workspace at any time to conduct a study, provided that they comply with all Data Use Policies and self-declare their research purpose. This information is regularly audited and made accessible publicly on the All of Us Research Projects Directory. This streamlined access model is guided by the principles that: participants are research partners and maintaining their privacy and data security is paramount; their data should be made as accessible as possible for authorized researchers; and we should continually seek to remove unnecessary barriers to accessing and using All of Us data.
For researchers at institutions with an existing institutional data use agreement, access can be gained as soon as they complete the required verification and compliance steps. As of August 2023, 556 institutions have agreements in place, allowing more than 5,000 approved researchers to actively work on more than 4,400 projects. The median time for a researcher from initial registration to completion of these requirements is 28.6 h (10th percentile: 48 min, 90th percentile: 14.9 days), a fraction of the weeks to months it can take to assemble a project-specific application and have it reviewed by an access board with conventional access models.
Given that the size of the project’s phenotypic and genomic dataset is expected to reach 4.75 PB in 2023, the use of a central data store and cloud analysis tools will save funders an estimated US$16.5 million per year when compared to the typical approach of allowing researchers to download genomic data. Storing one copy per institution of this data at 556 registered institutions would cost about US$1.16 billion per year. By contrast, storing a central cloud copy costs about US$1.14 million per year, a 99.9% saving. Importantly, cloud infrastructure also democratizes data access particularly for researchers who do not have high-performance local compute resources.
Here we present the All of Us Research Program’s approach to generating diverse clinical-grade genomic data at an unprecedented scale. We present the data release of about 245,000 genome sequences as part of a scalable framework that will grow to include genetic information and health data for one million or more people living across the USA. Our observations permit several conclusions.
First, the All of Us programme is making a notable contribution to improving the study of human biology through purposeful inclusion of under-represented individuals at scale 29 , 30 . Of the participants with genomic data in All of Us, 45.92% self-identified as a non-European race or ethnicity. This diversity enabled identification of more than 275 million new genetic variants across the dataset not previously captured by other large-scale genome aggregation efforts with diverse participants that have submitted variation to dbSNP v153, such as NHLBI TOPMed 31 freeze 8 (Extended Data Table 1 ). In contrast to gnomAD, All of Us permits individual-level genotype access with detailed phenotype data for all participants. Furthermore, unlike many genomics resources, All of Us is uniformly consented for general research use and enables researchers to go from initial account creation to individual-level data access in as little as a few hours. The All of Us cohort is significantly more diverse than those of other large contemporary research studies generating WGS data 32 , 33 . This enables a more equitable future for precision medicine (for example, through constructing polygenic risk scores that are appropriately calibrated to diverse populations 34 , 35 as the eMERGE programme has done leveraging All of Us data 36 , 37 ). Developing new tools and regulatory frameworks to enable analyses across multiple biobanks in the cloud to harness the unique strengths of each is an active area of investigation addressed in a companion paper to this work 38 .
Second, the All of Us Researcher Workbench embodies the programme’s design philosophy of open science, reproducible research, equitable access and transparency to researchers and to research participants 26 . Importantly, for research studies, no group of data users should have privileged access to All of Us resources based on anything other than data protection criteria. Although the All of Us Researcher Workbench initially targeted onboarding US academic, health care and non-profit organizations, it has recently expanded to international researchers. We anticipate further genomic and phenotypic data releases at regular intervals with data available to all researcher communities. We also anticipate additional derived data and functionality to be made available, such as reference data, structural variants and a service for array imputation using the All of Us genomic data.
Third, All of Us enables studying human biology at an unprecedented scale. The programmatic goal of sequencing one million or more genomes has required harnessing the output of multiple sequencing centres. Previous work has focused on achieving functional equivalence in data processing and joint calling pipelines 39 . To achieve clinical-grade data equivalence, All of Us required protocol equivalence at both sequencing production level and data processing across the sequencing centres. Furthermore, previous work has demonstrated the value of joint calling at scale 10 , 18 . The new GVS framework developed by the All of Us programme enables joint calling at extreme scales (Code availability). Finally, the provision of data access through cloud-native tools enables scalable and secure access and analysis to researchers while simultaneously enabling the trust of research participants and transparency underlying the All of Us data passport access model.
The clinical-grade sequencing carried out by All of Us enables not only research, but also the return of value to participants through clinically relevant genetic results and health-related traits to those who opt-in to receiving this information. In the years ahead, we anticipate that this partnership with All of Us participants will enable researchers to move beyond large-scale genomic discovery to understanding the consequences of implementing genomic medicine at scale.
The All of Us cohort
All of Us aims to engage a longitudinal cohort of one million or more US participants, with a focus on including populations that have historically been under-represented in biomedical research. Details of the All of Us cohort have been described previously 5 . Briefly, the primary objective is to build a robust research resource that can facilitate the exploration of biological, clinical, social and environmental determinants of health and disease. The programme will collect and curate health-related data and biospecimens, and these data and biospecimens will be made broadly available for research uses. Health data are obtained through the electronic medical record and through participant surveys. Survey templates can be found on our public website: https://www.researchallofus.org/data-tools/survey-explorer/ . Adults 18 years and older who have the capacity to consent and reside in the USA or a US territory at present are eligible. Informed consent for all participants is conducted in person or through an eConsent platform that includes primary consent, HIPAA Authorization for Research use of EHRs and other external health data, and Consent for Return of Genomic Results. The protocol was reviewed by the Institutional Review Board (IRB) of the All of Us Research Program. The All of Us IRB follows the regulations and guidance of the NIH Office for Human Research Protections for all studies, ensuring that the rights and welfare of research participants are overseen and protected uniformly.
Data accessibility through a ‘data passport’
Authorization for access to participant-level data in All of Us is based on a ‘data passport’ model, through which authorized researchers do not need IRB review for each research project. The data passport is required for gaining data access to the Researcher Workbench and for creating workspaces to carry out research projects using All of Us data. At present, data passports are authorized through a six-step process that includes affiliation with an institution that has signed a Data Use and Registration Agreement, account creation, identity verification, completion of ethics training, and attestation to a data user code of conduct. Results reported follow the All of Us Data and Statistics Dissemination Policy disallowing disclosure of group counts under 20 to protect participant privacy without seeking prior approval 40 .
At present, All of Us gathers EHR data from about 50 health care organizations that are funded to recruit and enrol participants as well as transfer EHR data for those participants who have consented to provide them. Data stewards at each provider organization harmonize their local data to the Observational Medical Outcomes Partnership (OMOP) Common Data Model, and then submit it to the All of Us Data and Research Center (DRC) so that it can be linked with other participant data and further curated for research use. OMOP is a common data model standardizing health information from disparate EHRs to common vocabularies and organized into tables according to data domains. EHR data are updated from the recruitment sites and sent to the DRC quarterly. Updated data releases to the research community occur approximately once a year. Supplementary Table 6 outlines the OMOP concepts collected by the DRC quarterly from the recruitment sites.
Biospecimen collection and processing
Participants who consented to participate in All of Us donated fresh whole blood (4 ml EDTA and 10 ml EDTA) as a primary source of DNA. The All of Us Biobank managed by the Mayo Clinic extracted DNA from 4 ml EDTA whole blood, and DNA was stored at −80 °C at an average concentration of 150 ng µl −1 . The buffy coat isolated from 10 ml EDTA whole blood has been used for extracting DNA in the case of initial extraction failure or absence of 4 ml EDTA whole blood. The Biobank plated 2.4 µg DNA with a concentration of 60 ng µl −1 in duplicate for array and WGS samples. The samples are distributed to All of Us Genome Centers weekly, and a negative (empty well) control and National Institute of Standards and Technology controls are incorporated every two months for QC purposes.
Genome Center sample receipt, accession and QC
On receipt of DNA sample shipments, the All of Us Genome Centers carry out an inspection of the packaging and sample containers to ensure that sample integrity has not been compromised during transport and to verify that the sample containers correspond to the shipping manifest. QC of the submitted samples also includes DNA quantification, using routine procedures to confirm volume and concentration (Supplementary Table 7 ). Any issues or discrepancies are recorded, and affected samples are put on hold until resolved. Samples that meet quality thresholds are accessioned in the Laboratory Information Management System, and sample aliquots are prepared for library construction processing (for example, normalized with respect to concentration and volume).
WGS library construction, sequencing and primary data QC
The DNA sample is first sheared using a Covaris sonicator and is then size-selected using AMPure XP beads to restrict the range of library insert sizes. Using the PCR Free Kapa HyperPrep library construction kit, enzymatic steps are completed to repair the jagged ends of DNA fragments, add proper A-base segments, and ligate indexed adapter barcode sequences onto samples. Excess adaptors are removed using AMPure XP beads for a final clean-up. Libraries are quantified using quantitative PCR with the Illumina Kapa DNA Quantification Kit and then normalized and pooled for sequencing (Supplementary Table 7 ).
Pooled libraries are loaded on the Illumina NovaSeq 6000 instrument. The data from the initial sequencing run are used to QC individual libraries and to remove non-conforming samples from the pipeline. The data are also used to calibrate the pooling volume of each individual library and re-pool the libraries for additional NovaSeq sequencing to reach an average coverage of 30×.
After demultiplexing, WGS analysis occurs on the Illumina DRAGEN platform. The DRAGEN pipeline consists of highly optimized algorithms for mapping, aligning, sorting, duplicate marking and haplotype variant calling and makes use of platform features such as compression and BCL conversion. Alignment uses the GRCh38dh reference genome. QC data are collected at every stage of the analysis protocol, providing high-resolution metrics required to ensure data consistency for large-scale multiplexing. The DRAGEN pipeline produces a large number of metrics that cover lane, library, flow cell, barcode and sample-level metrics for all runs as well as assessing contamination and mapping quality. The All of Us Genome Centers use these metrics to determine pass or fail for each sample before submitting the CRAM files to the All of Us DRC. For mapping and variant calling, all Genome Centers have harmonized on a set of DRAGEN parameters, which ensures consistency in processing (Supplementary Table 2 ).
Every step through the WGS procedure is rigorously controlled by predefined QC measures. Various control mechanisms and acceptance criteria were established during WGS assay validation. Specific metrics for reviewing and releasing genome data are: mean coverage (threshold of ≥30×), genome coverage (threshold of ≥90% at 20×), coverage of hereditary disease risk genes (threshold of ≥95% at 20×), aligned Q30 bases (threshold of ≥8 × 10 10 ), contamination (threshold of ≤1%) and concordance to independently processed array data.
Array genotyping
Samples are processed for genotyping at three All of Us Genome Centers (Broad, Johns Hopkins University and University of Washington). DNA samples are received from the Biobank and the process is facilitated by the All of Us genomics workflow described above. All three centres used an identical array product, scanners, resource files and genotype calling software for array processing to reduce batch effects. Each centre has its own Laboratory Information Management System that manages workflow control, sample and reagent tracking, and centre-specific liquid handling robotics.
Samples are processed using the Illumina Global Diversity Array (GDA) with Illumina Infinium LCG chemistry using the automated protocol and scanned on Illumina iSCANs with Automated Array Loaders. Illumina IAAP software converts raw data (IDAT files; 2 per sample) into a single GTC file per sample using the BPM file (defines strand, probe sequences and illumicode address) and the EGT file (defines the relationship between intensities and genotype calls). Files used for this data release are: GDA-8v1-0_A5.bpm, GDA-8v1-0_A1_ClusterFile.egt, gentrain v3, reference hg19 and gencall cutoff 0.15. The GDA array assays a total of 1,914,935 variant positions including 1,790,654 single-nucleotide variants, 44,172 indels, 9,935 intensity-only probes for CNV calling, and 70,174 duplicates (same position, different probes). Picard GtcToVcf is used to convert the GTC files to VCF format. Resulting VCF and IDAT files are submitted to the DRC for ingestion and further processing. The VCF file contains assay name, chromosome, position, genotype calls, quality score, raw and normalized intensities, B allele frequency and log R ratio values. Each genome centre is running the GDA array under Clinical Laboratory Improvement Amendments-compliant protocols. The GTC files are parsed and metrics are uploaded to in-house Laboratory Information Management System systems for QC review.
At batch level (each set of 96-well plates run together in the laboratory at one time), each genome centre includes positive control samples that are required to have >98% call rate and >99% concordance to existing data to approve release of the batch of data. At the sample level, the call rate and sex are the key QC determinants 41 . Contamination is also measured using BAFRegress 42 and reported out as metadata. Any sample with a call rate below 98% is repeated one time in the laboratory. Genotyped sex is determined by plotting normalized x versus normalized y intensity values for a batch of samples. Any sample discordant with ‘sex at birth’ reported by the All of Us participant is flagged for further detailed review and repeated one time in the laboratory. If several sex-discordant samples are clustered on an array or on a 96-well plate, the entire array or plate will have data production repeated. Samples identified with sex chromosome aneuploidies are also reported back as metadata (XXX, XXY, XYY and so on). A final processing status of ‘pass’, ‘fail’ or ‘abandon’ is determined before release of data to the All of Us DRC. An array sample will pass if the call rate is >98% and the genotyped sex and sex at birth are concordant (or the sex at birth is not applicable). An array sample will fail if the genotyped sex and the sex at birth are discordant. An array sample will have the status of abandon if the call rate is <98% after at least two attempts at the genome centre.
Data from the arrays are used for participant return of genetic ancestry and non-health-related traits for those who consent, and they are also used to facilitate additional QC of the matched WGS data. Contamination is assessed in the array data to determine whether DNA re-extraction is required before WGS. Re-extraction is prompted by level of contamination combined with consent status for return of results. The arrays are also used to confirm sample identity between the WGS data and the matched array data by assessing concordance at 100 unique sites. To establish concordance, a fingerprint file of these 100 sites is provided to the Genome Centers to assess concordance with the same sites in the WGS data before CRAM submission.
Genomic data curation
As seen in Extended Data Fig. 2 , we generate a joint call set for all WGS samples and make these data available in their entirety and by sample subsets to researchers. A breakdown of the frequencies, stratified by computed ancestries for which we had more than 10,000 participants can be found in Extended Data Fig. 3 . The joint call set process allows us to leverage information across samples to improve QC and increase accuracy.
Single-sample QC
If a sample fails single-sample QC, it is excluded from the release and is not reported in this document. These tests detect sample swaps, cross-individual contamination and sample preparation errors. In some cases, we carry out these tests twice (at both the Genome Center and the DRC), for two reasons: to confirm internal consistency between sites; and to mark samples as passing (or failing) QC on the basis of the research pipeline criteria. The single-sample QC process accepts a higher contamination rate than the clinical pipeline (0.03 for the research pipeline versus 0.01 for the clinical pipeline), but otherwise uses identical thresholds. The list of specific QC processes, passing criteria, error modes addressed and an overview of the results can be found in Supplementary Table 3 .
Joint call set QC
During joint calling, we carry out additional QC steps using information that is available across samples including hard thresholds, population outliers, allele-specific filters, and sensitivity and precision evaluation. Supplementary Table 4 summarizes both the steps that we took and the results obtained for the WGS data. More detailed information about the methods and specific parameters can be found in the All of Us Genomic Research Data Quality Report 36 .
Batch effect analysis
We analysed cross-sequencing centre batch effects in the joint call set. To quantify the batch effect, we calculated Cohen’s d (ref. 43 ) for four metrics (insertion/deletion ratio, single-nucleotide polymorphism count, indel count and single-nucleotide polymorphism transition/transversion ratio) across the three genome sequencing centres (Baylor College of Medicine, Broad Institute and University of Washington), stratified by computed ancestry and seven regions of the genome (whole genome, high-confidence calling, repetitive, GC content of >0.85, GC content of <0.15, low mappability, the ACMG59 genes and regions of large duplications (>1 kb)). Using random batches as a control set, all comparisons had a Cohen’s d of <0.35. Here we report any Cohen’s d results >0.5, which we chose before this analysis and is conventionally the threshold of a medium effect size 44 .
We found that there was an effect size in indel counts (Cohen’s d of 0.53) in the entire genome, between Broad Institute and University of Washington, but this was being driven by repetitive and low-mappability regions. We found no batch effects with Cohen’s d of >0.5 in the ratio metrics or in any metrics in the high-confidence calling, low or high GC content, or ACMG59 regions. A complete list of the batch effects with Cohen’s d of >0.5 are found in Supplementary Table 8 .
Sensitivity and precision evaluation
To determine sensitivity and precision, we included four well-characterized control samples (four National Institute of Standards and Technology Genome in a Bottle samples (HG-001, HG-003, HG-004 and HG-005). The samples were sequenced with the same protocol as All of Us. Of note, these samples were not included in data released to researchers. We used the corresponding published set of variant calls for each sample as the ground truth in our sensitivity and precision calculations. We use the high-confidence calling region, defined by Genome in a Bottle v4.2.1, as the source of ground truth. To be called a true positive, a variant must match the chromosome, position, reference allele, alternate allele and zygosity. In cases of sites with multiple alternative alleles, each alternative allele is considered separately. Sensitivity and precision results are reported in Supplementary Table 5 .
Genetic ancestry inference
We computed categorical ancestry for all WGS samples in All of Us and made these available to researchers. These predictions are also the basis for population allele frequency calculations in the Genomic Variants section of the public Data Browser. We used the high-quality set of sites to determine an ancestry label for each sample. The ancestry categories are based on the same labels used in gnomAD 18 , the Human Genome Diversity Project (HGDP) 45 and 1000 Genomes 1 : African (AFR); Latino/admixed American (AMR); East Asian (EAS); Middle Eastern (MID); European (EUR), composed of Finnish (FIN) and Non-Finnish European (NFE); Other (OTH), not belonging to one of the other ancestries or is an admixture; South Asian (SAS).
We trained a random forest classifier 46 on a training set of the HGDP and 1000 Genomes samples variants on the autosome, obtained from gnomAD 11 . We generated the first 16 principal components (PCs) of the training sample genotypes (using the hwe_normalized_pca in Hail) at the high-quality variant sites for use as the feature vector for each training sample. We used the truth labels from the sample metadata, which can be found alongside the VCFs. Note that we do not train the classifier on the samples labelled as Other. We use the label probabilities (‘confidence’) of the classifier on the other ancestries to determine ancestry of Other.
To determine the ancestry of All of Us samples, we project the All of Us samples into the PCA space of the training data and apply the classifier. As a proxy for the accuracy of our All of Us predictions, we look at the concordance between the survey results and the predicted ancestry. The concordance between self-reported ethnicity and the ancestry predictions was 87.7%.
PC data from All of Us samples and the HGDP and 1000 Genomes samples were used to compute individual participant genetic ancestry fractions for All of Us samples using the Rye program. Rye uses PC data to carry out rapid and accurate genetic ancestry inference on biobank-scale datasets 47 . HGDP and 1000 Genomes reference samples were used to define a set of six distinct and coherent ancestry groups—African, East Asian, European, Middle Eastern, Latino/admixed American and South Asian—corresponding to participant self-identified race and ethnicity groups. Rye was run on the first 16 PCs, using the defined reference ancestry groups to assign ancestry group fractions to individual All of Us participant samples.

Relatedness
We calculated the kinship score using the Hail pc_relate function and reported any pairs with a kinship score above 0.1. The kinship score is half of the fraction of the genetic material shared (ranges from 0.0 to 0.5). We determined the maximal independent set 41 for related samples. We identified a maximally unrelated set of 231,442 samples (94%) for kinship scored greater than 0.1.
LDL-C common variant GWAS
The phenotypic data were extracted from the Curated Data Repository (CDR, Control Tier Dataset v7) in the All of Us Researcher Workbench. The All of Us Cohort Builder and Dataset Builder were used to extract all LDL cholesterol measurements from the Lab and Measurements criteria in EHR data for all participants who have WGS data. The most recent measurements were selected as the phenotype and adjusted for statin use 19 , age and sex. A rank-based inverse normal transformation was applied for this continuous trait to increase power and deflate type I error. Analysis was carried out on the Hail MatrixTable representation of the All of Us WGS joint-called data including removing monomorphic variants, variants with a call rate of <95% and variants with extreme Hardy–Weinberg equilibrium values ( P < 10 −15 ). A linear regression was carried out with REGENIE 48 on variants with a minor allele frequency >5%, further adjusting for relatedness to the first five ancestry PCs. The final analysis included 34,924 participants and 8,589,520 variants.
Genotype-by-phenotype replication
We tested replication rates of known phenotype–genotype associations in three of the four largest populations: EUR, AFR and EAS. The AMR population was not included because they have no registered GWAS. This method is a conceptual extension of the original GWAS × phenome-wide association study, which replicated 66% of powered associations in a single EHR-linked biobank 49 . The PGRM is an expansion of this work by Bastarache et al., based on associations in the GWAS catalogue 50 in June 2020 (ref. 51 ). After directly matching the Experimental Factor Ontology terms to phecodes, the authors identified 8,085 unique loci and 170 unique phecodes that compose the PGRM. They showed replication rates in several EHR-linked biobanks ranging from 76% to 85%. For this analysis, we used the EUR-, and AFR-based maps, considering only catalogue associations that were P < 5 × 10 −8 significant.
The main tools used were the Python package Hail for data extraction, plink for genomic associations, and the R packages PheWAS and pgrm for further analysis and visualization. The phenotypes, participant-reported sex at birth, and year of birth were extracted from the All of Us CDR (Controlled Tier Dataset v7). These phenotypes were then loaded into a plink-compatible format using the PheWAS package, and related samples were removed by sub-setting to the maximally unrelated dataset ( n = 231,442). Only samples with EHR data were kept, filtered by selected loci, annotated with demographic and phenotypic information extracted from the CDR and ancestry prediction information provided by All of Us, ultimately resulting in 181,345 participants for downstream analysis. The variants in the PGRM were filtered by a minimum population-specific allele frequency of >1% or population-specific allele count of >100, leaving 4,986 variants. Results for which there were at least 20 cases in the ancestry group were included. Then, a series of Firth logistic regression tests with phecodes as the outcome and variants as the predictor were carried out, adjusting for age, sex (for non-sex-specific phenotypes) and the first three genomic PC features as covariates. The PGRM was annotated with power calculations based on the case counts and reported allele frequencies. Power of 80% or greater was considered powered for this analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The All of Us Research Hub has a tiered data access data passport model with three data access tiers. The Public Tier dataset contains only aggregate data with identifiers removed. These data are available to the public through Data Snapshots ( https://www.researchallofus.org/data-tools/data-snapshots/ ) and the public Data Browser ( https://databrowser.researchallofus.org/ ). The Registered Tier curated dataset contains individual-level data, available only to approved researchers on the Researcher Workbench. At present, the Registered Tier includes data from EHRs, wearables and surveys, as well as physical measurements taken at the time of participant enrolment. The Controlled Tier dataset contains all data in the Registered Tier and additionally genomic data in the form of WGS and genotyping arrays, previously suppressed demographic data fields from EHRs and surveys, and unshifted dates of events. At present, Registered Tier and Controlled Tier data are available to researchers at academic institutions, non-profit institutions, and both non-profit and for-profit health care institutions. Work is underway to begin extending access to additional audiences, including industry-affiliated researchers. Researchers have the option to register for Registered Tier and/or Controlled Tier access by completing the All of Us Researcher Workbench access process, which includes identity verification and All of Us-specific training in research involving human participants ( https://www.researchallofus.org/register/ ). Researchers may create a new workspace at any time to conduct any research study, provided that they comply with all Data Use Policies and self-declare their research purpose. This information is made accessible publicly on the All of Us Research Projects Directory at https://allofus.nih.gov/protecting-data-and-privacy/research-projects-all-us-data .
Code availability
The GVS code is available at https://github.com/broadinstitute/gatk/tree/ah_var_store/scripts/variantstore . The LDL GWAS pipeline is available as a demonstration project in the Featured Workspace Library on the Researcher Workbench ( https://workbench.researchallofus.org/workspaces/aou-rw-5981f9dc/aouldlgwasregeniedsubctv6duplicate/notebooks ).
The 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526 , 68–74 (2015).
Article Google Scholar
Claussnitzer, M. et al. A brief history of human disease genetics. Nature 577 , 179–189 (2020).
Article CAS PubMed PubMed Central ADS Google Scholar
Wojcik, G. L. et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature 570 , 514–518 (2019).
Article CAS PubMed PubMed Central Google Scholar
Lewis, A. C. F. et al. Getting genetic ancestry right for science and society. Science 376 , 250–252 (2022).
All of Us Program Investigators. The “All of Us” Research Program. N. Engl. J. Med. 381 , 668–676 (2019).
Ramirez, A. H., Gebo, K. A. & Harris, P. A. Progress with the All of Us Research Program: opening access for researchers. JAMA 325 , 2441–2442 (2021).
Article PubMed Google Scholar
Ramirez, A. H. et al. The All of Us Research Program: data quality, utility, and diversity. Patterns 3 , 100570 (2022).
Article PubMed PubMed Central Google Scholar
Overhage, J. M., Ryan, P. B., Reich, C. G., Hartzema, A. G. & Stang, P. E. Validation of a common data model for active safety surveillance research. J. Am. Med. Inform. Assoc. 19 , 54–60 (2012).
Venner, E. et al. Whole-genome sequencing as an investigational device for return of hereditary disease risk and pharmacogenomic results as part of the All of Us Research Program. Genome Med. 14 , 34 (2022).
Lek, M. et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536 , 285–291 (2016).
Tiao, G. & Goodrich, J. gnomAD v3.1 New Content, Methods, Annotations, and Data Availability ; https://gnomad.broadinstitute.org/news/2020-10-gnomad-v3-1-new-content-methods-annotations-and-data-availability/ .
Chen, S. et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature 625 , 92–100 (2022).
Zook, J. M. et al. An open resource for accurately benchmarking small variant and reference calls. Nat. Biotechnol. 37 , 561–566 (2019).
Krusche, P. et al. Best practices for benchmarking germline small-variant calls in human genomes. Nat. Biotechnol. 37 , 555–560 (2019).
Stromberg, M. et al. Nirvana: clinical grade variant annotator. In Proc. 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 596 (Association for Computing Machinery, 2017).
Sherry, S. T. et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29 , 308–311 (2001).
Venner, E. et al. The frequency of pathogenic variation in the All of Us cohort reveals ancestry-driven disparities. Commun. Biol. https://doi.org/10.1038/s42003-023-05708-y (2024).
Karczewski, S. et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581 , 434–443 (2020).
Selvaraj, M. S. et al. Whole genome sequence analysis of blood lipid levels in >66,000 individuals. Nat. Commun. 13 , 5995 (2022).
Wang, X. et al. Common and rare variants associated with cardiometabolic traits across 98,622 whole-genome sequences in the All of Us research program. J. Hum. Genet. 68 , 565–570 (2023).
Bastarache, L. et al. The phenotype-genotype reference map: improving biobank data science through replication. Am. J. Hum. Genet. 110 , 1522–1533 (2023).
Bianchi, D. W. et al. The All of Us Research Program is an opportunity to enhance the diversity of US biomedical research. Nat. Med. https://doi.org/10.1038/s41591-023-02744-3 (2024).
Van Driest, S. L. et al. Association between a common, benign genotype and unnecessary bone marrow biopsies among African American patients. JAMA Intern. Med. 181 , 1100–1105 (2021).
Chen, M.-H. et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell 182 , 1198–1213 (2020).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594 , 398–402 (2021).
Hu, X. et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat. Genet. 47 , 898–905 (2015).
Grant, S. F. A. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat. Genet. 38 , 320–323 (2006).
Article CAS PubMed Google Scholar
All of Us Research Program. Framework for Access to All of Us Data Resources v1.1 (2021); https://www.researchallofus.org/wp-content/themes/research-hub-wordpress-theme/media/data&tools/data-access-use/AoU_Data_Access_Framework_508.pdf .
Abul-Husn, N. S. & Kenny, E. E. Personalized medicine and the power of electronic health records. Cell 177 , 58–69 (2019).
Mapes, B. M. et al. Diversity and inclusion for the All of Us research program: A scoping review. PLoS ONE 15 , e0234962 (2020).
Taliun, D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590 , 290–299 (2021).
Bycroft, C. et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562 , 203–209 (2018).
Halldorsson, B. V. et al. The sequences of 150,119 genomes in the UK Biobank. Nature 607 , 732–740 (2022).
Kurniansyah, N. et al. Evaluating the use of blood pressure polygenic risk scores across race/ethnic background groups. Nat. Commun. 14 , 3202 (2023).
Hou, K. et al. Causal effects on complex traits are similar for common variants across segments of different continental ancestries within admixed individuals. Nat. Genet. 55 , 549– 558 (2022).
Linder, J. E. et al. Returning integrated genomic risk and clinical recommendations: the eMERGE study. Genet. Med. 25 , 100006 (2023).
Lennon, N. J. et al. Selection, optimization and validation of ten chronic disease polygenic risk scores for clinical implementation in diverse US populations. Nat. Med. https://doi.org/10.1038/s41591-024-02796-z (2024).
Deflaux, N. et al. Demonstrating paths for unlocking the value of cloud genomics through cross cohort analysis. Nat. Commun. 14 , 5419 (2023).
Regier, A. A. et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat. Commun. 9 , 4038 (2018).
Article PubMed PubMed Central ADS Google Scholar
All of Us Research Program. Data and Statistics Dissemination Policy (2020); https://www.researchallofus.org/wp-content/themes/research-hub-wordpress-theme/media/2020/05/AoU_Policy_Data_and_Statistics_Dissemination_508.pdf .
Laurie, C. C. et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet. Epidemiol. 34 , 591–602 (2010).
Jun, G. et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am. J. Hum. Genet. 91 , 839–848 (2012).
Cohen, J. Statistical Power Analysis for the Behavioral Sciences (Routledge, 2013).
Andrade, C. Mean difference, standardized mean difference (SMD), and their use in meta-analysis. J. Clin. Psychiatry 81 , 20f13681 (2020).
Cavalli-Sforza, L. L. The Human Genome Diversity Project: past, present and future. Nat. Rev. Genet. 6 , 333–340 (2005).
Ho, T. K. Random decision forests. In Proc. 3rd International Conference on Document Analysis and Recognition (IEEE Computer Society Press, 2002).
Conley, A. B. et al. Rye: genetic ancestry inference at biobank scale. Nucleic Acids Res. 51 , e44 (2023).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53 , 1097–1103 (2021).
Denny, J. C. Systematic comparison of phenome-wide association study of electronic medical record data and genome-wide association study data. Nat. Biotech. 31 , 1102–1111 (2013).
Buniello, A. et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47 , D1005–D1012 (2019).
Bastarache, L. et al. The Phenotype-Genotype Reference Map: improving biobank data science through replication. Am. J. Hum. Genet. 10 , 1522–1533 (2023).
Download references
Acknowledgements
The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers (OT2 OD026549; OT2 OD026554; OT2 OD026557; OT2 OD026556; OT2 OD026550; OT2 OD 026552; OT2 OD026553; OT2 OD026548; OT2 OD026551; OT2 OD026555); Inter agency agreement AOD 16037; Federally Qualified Health Centers HHSN 263201600085U; Data and Research Center: U2C OD023196; Genome Centers (OT2 OD002748; OT2 OD002750; OT2 OD002751); Biobank: U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: U24 OD023163; Communications and Engagement: OT2 OD023205; OT2 OD023206; and Community Partners (OT2 OD025277; OT2 OD025315; OT2 OD025337; OT2 OD025276). In addition, the All of Us Research Program would not be possible without the partnership of its participants. All of Us and the All of Us logo are service marks of the US Department of Health and Human Services. E.E.E. is an investigator of the Howard Hughes Medical Institute. We acknowledge the foundational contributions of our friend and colleague, the late Deborah A. Nickerson. Debbie’s years of insightful contributions throughout the formation of the All of Us genomics programme are permanently imprinted, and she shares credit for all of the successes of this programme.
Author information
Authors and affiliations.
Division of Genetic Medicine, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA
Alexander G. Bick & Henry R. Condon
Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX, USA
Ginger A. Metcalf, Eric Boerwinkle, Richard A. Gibbs, Donna M. Muzny, Eric Venner, Kimberly Walker, Jianhong Hu, Harsha Doddapaneni, Christie L. Kovar, Mullai Murugan, Shannon Dugan, Ziad Khan & Eric Boerwinkle
Vanderbilt Institute of Clinical and Translational Research, Vanderbilt University Medical Center, Nashville, TN, USA
Kelsey R. Mayo, Jodell E. Linder, Melissa Basford, Ashley Able, Ashley E. Green, Robert J. Carroll, Jennifer Zhang & Yuanyuan Wang
Data Sciences Platform, Broad Institute of MIT and Harvard, Cambridge, MA, USA
Lee Lichtenstein, Anthony Philippakis, Sophie Schwartz, M. Morgan T. Aster, Kristian Cibulskis, Andrea Haessly, Rebecca Asch, Aurora Cremer, Kylee Degatano, Akum Shergill, Laura D. Gauthier, Samuel K. Lee, Aaron Hatcher, George B. Grant, Genevieve R. Brandt, Miguel Covarrubias, Eric Banks & Wail Baalawi
Verily, South San Francisco, CA, USA
Shimon Rura, David Glazer, Moira K. Dillon & C. H. Albach
Department of Biomedical Informatics, Vanderbilt University Medical Center, Nashville, TN, USA
Robert J. Carroll, Paul A. Harris & Dan M. Roden
All of Us Research Program, National Institutes of Health, Bethesda, MD, USA
Anjene Musick, Andrea H. Ramirez, Sokny Lim, Siddhartha Nambiar, Bradley Ozenberger, Anastasia L. Wise, Chris Lunt, Geoffrey S. Ginsburg & Joshua C. Denny
School of Biological Sciences, Georgia Institute of Technology, Atlanta, GA, USA
I. King Jordan, Shashwat Deepali Nagar & Shivam Sharma
Neuroscience Institute, Institute of Translational Genomic Medicine, Morehouse School of Medicine, Atlanta, GA, USA
Robert Meller
Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA
Mine S. Cicek, Stephen N. Thibodeau & Mine S. Cicek
Department of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, MD, USA
Kimberly F. Doheny, Michelle Z. Mawhinney, Sean M. L. Griffith, Elvin Hsu, Hua Ling & Marcia K. Adams
Department of Genome Sciences, University of Washington School of Medicine, Seattle, WA, USA
Evan E. Eichler, Joshua D. Smith, Christian D. Frazar, Colleen P. Davis, Karynne E. Patterson, Marsha M. Wheeler, Sean McGee, Mitzi L. Murray, Valeria Vasta, Dru Leistritz, Matthew A. Richardson, Aparna Radhakrishnan & Brenna W. Ehmen
Howard Hughes Medical Institute, University of Washington, Seattle, WA, USA
Evan E. Eichler
Broad Institute of MIT and Harvard, Cambridge, MA, USA
Stacey Gabriel, Heidi L. Rehm, Niall J. Lennon, Christina Austin-Tse, Eric Banks, Michael Gatzen, Namrata Gupta, Katie Larsson, Sheli McDonough, Steven M. Harrison, Christopher Kachulis, Matthew S. Lebo, Seung Hoan Choi & Xin Wang
Division of Medical Genetics, Department of Medicine, University of Washington School of Medicine, Seattle, WA, USA
Gail P. Jarvik & Elisabeth A. Rosenthal
Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA
Dan M. Roden
Department of Pharmacology, Vanderbilt University Medical Center, Nashville, TN, USA
Center for Individualized Medicine, Biorepository Program, Mayo Clinic, Rochester, MN, USA
Stephen N. Thibodeau, Ashley L. Blegen, Samantha J. Wirkus, Victoria A. Wagner, Jeffrey G. Meyer & Mine S. Cicek
Color Health, Burlingame, CA, USA
Scott Topper, Cynthia L. Neben, Marcie Steeves & Alicia Y. Zhou
School of Public Health, University of Texas Health Science Center at Houston, Houston, TX, USA
Eric Boerwinkle
Laboratory for Molecular Medicine, Massachusetts General Brigham Personalized Medicine, Cambridge, MA, USA
Christina Austin-Tse, Emma Henricks & Matthew S. Lebo
Department of Laboratory Medicine and Pathology, University of Washington School of Medicine, Seattle, WA, USA
Christina M. Lockwood, Brian H. Shirts, Colin C. Pritchard, Jillian G. Buchan & Niklas Krumm
Manuscript Writing Group
- Alexander G. Bick
- , Ginger A. Metcalf
- , Kelsey R. Mayo
- , Lee Lichtenstein
- , Shimon Rura
- , Robert J. Carroll
- , Anjene Musick
- , Jodell E. Linder
- , I. King Jordan
- , Shashwat Deepali Nagar
- , Shivam Sharma
- & Robert Meller
All of Us Research Program Genomics Principal Investigators
- Melissa Basford
- , Eric Boerwinkle
- , Mine S. Cicek
- , Kimberly F. Doheny
- , Evan E. Eichler
- , Stacey Gabriel
- , Richard A. Gibbs
- , David Glazer
- , Paul A. Harris
- , Gail P. Jarvik
- , Anthony Philippakis
- , Heidi L. Rehm
- , Dan M. Roden
- , Stephen N. Thibodeau
- & Scott Topper
Biobank, Mayo
- Ashley L. Blegen
- , Samantha J. Wirkus
- , Victoria A. Wagner
- , Jeffrey G. Meyer
- & Stephen N. Thibodeau
Genome Center: Baylor-Hopkins Clinical Genome Center
- Donna M. Muzny
- , Eric Venner
- , Michelle Z. Mawhinney
- , Sean M. L. Griffith
- , Elvin Hsu
- , Marcia K. Adams
- , Kimberly Walker
- , Jianhong Hu
- , Harsha Doddapaneni
- , Christie L. Kovar
- , Mullai Murugan
- , Shannon Dugan
- , Ziad Khan
- & Richard A. Gibbs
Genome Center: Broad, Color, and Mass General Brigham Laboratory for Molecular Medicine
- Niall J. Lennon
- , Christina Austin-Tse
- , Eric Banks
- , Michael Gatzen
- , Namrata Gupta
- , Emma Henricks
- , Katie Larsson
- , Sheli McDonough
- , Steven M. Harrison
- , Christopher Kachulis
- , Matthew S. Lebo
- , Cynthia L. Neben
- , Marcie Steeves
- , Alicia Y. Zhou
- , Scott Topper
- & Stacey Gabriel
Genome Center: University of Washington
- Gail P. Jarvik
- , Joshua D. Smith
- , Christian D. Frazar
- , Colleen P. Davis
- , Karynne E. Patterson
- , Marsha M. Wheeler
- , Sean McGee
- , Christina M. Lockwood
- , Brian H. Shirts
- , Colin C. Pritchard
- , Mitzi L. Murray
- , Valeria Vasta
- , Dru Leistritz
- , Matthew A. Richardson
- , Jillian G. Buchan
- , Aparna Radhakrishnan
- , Niklas Krumm
- & Brenna W. Ehmen
Data and Research Center
- Lee Lichtenstein
- , Sophie Schwartz
- , M. Morgan T. Aster
- , Kristian Cibulskis
- , Andrea Haessly
- , Rebecca Asch
- , Aurora Cremer
- , Kylee Degatano
- , Akum Shergill
- , Laura D. Gauthier
- , Samuel K. Lee
- , Aaron Hatcher
- , George B. Grant
- , Genevieve R. Brandt
- , Miguel Covarrubias
- , Melissa Basford
- , Alexander G. Bick
- , Ashley Able
- , Ashley E. Green
- , Jennifer Zhang
- , Henry R. Condon
- , Yuanyuan Wang
- , Moira K. Dillon
- , C. H. Albach
- , Wail Baalawi
- & Dan M. Roden
All of Us Research Demonstration Project Teams
- Seung Hoan Choi
- , Elisabeth A. Rosenthal
NIH All of Us Research Program Staff
- Andrea H. Ramirez
- , Sokny Lim
- , Siddhartha Nambiar
- , Bradley Ozenberger
- , Anastasia L. Wise
- , Chris Lunt
- , Geoffrey S. Ginsburg
- & Joshua C. Denny
Contributions
The All of Us Biobank (Mayo Clinic) collected, stored and plated participant biospecimens. The All of Us Genome Centers (Baylor-Hopkins Clinical Genome Center; Broad, Color, and Mass General Brigham Laboratory for Molecular Medicine; and University of Washington School of Medicine) generated and QCed the whole-genomic data. The All of Us Data and Research Center (Vanderbilt University Medical Center, Broad Institute of MIT and Harvard, and Verily) generated the WGS joint call set, carried out quality assurance and QC analyses and developed the Researcher Workbench. All of Us Research Demonstration Project Teams contributed analyses. The other All of Us Genomics Investigators and NIH All of Us Research Program Staff provided crucial programmatic support. Members of the manuscript writing group (A.G.B., G.A.M., K.R.M., L.L., S.R., R.J.C. and A.M.) wrote the first draft of this manuscript, which was revised with contributions and feedback from all authors.
Corresponding author
Correspondence to Alexander G. Bick .
Ethics declarations
Competing interests.
D.M.M., G.A.M., E.V., K.W., J.H., H.D., C.L.K., M.M., S.D., Z.K., E. Boerwinkle and R.A.G. declare that Baylor Genetics is a Baylor College of Medicine affiliate that derives revenue from genetic testing. Eric Venner is affiliated with Codified Genomics, a provider of genetic interpretation. E.E.E. is a scientific advisory board member of Variant Bio, Inc. A.G.B. is a scientific advisory board member of TenSixteen Bio. The remaining authors declare no competing interests.
Peer review
Peer review information.
Nature thanks Timothy Frayling and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended data fig. 1 historic availability of ehr records in all of us v7 controlled tier curated data repository (n = 413,457)..
For better visibility, the plot shows growth starting in 2010.
Extended Data Fig. 2 Overview of the Genomic Data Curation Pipeline for WGS samples.
The Data and Research Center (DRC) performs additional single sample quality control (QC) on the data as it arrives from the Genome Centers. The variants from samples that pass this QC are loaded into the Genomic Variant Store (GVS), where we jointly call the variants and apply additional QC. We apply a joint call set QC process, which is stored with the call set. The entire joint call set is rendered as a Hail Variant Dataset (VDS), which can be accessed from the analysis notebooks in the Researcher Workbench. Subsections of the genome are extracted from the VDS and rendered in different formats with all participants. Auxiliary data can also be accessed through the Researcher Workbench. This includes variant functional annotations, joint call set QC results, predicted ancestry, and relatedness. Auxiliary data are derived from GVS (arrow not shown) and the VDS. The Cohort Builder directly queries GVS when researchers request genomic data for subsets of samples. Aligned reads, as cram files, are available in the Researcher Workbench (not shown). The graphics of the dish, gene and computer and the All of Us logo are reproduced with permission of the National Institutes of Health’s All of Us Research Program.
Extended Data Fig. 3 Proportion of allelic frequencies (AF), stratified by computed ancestry with over 10,000 participants.
Bar counts are not cumulative (eg, “pop AF < 0.01” does not include “pop AF < 0.001”).
Extended Data Fig. 4 Distribution of pathogenic, and likely pathogenic ClinVar variants.
Stratified by ancestry filtered to only those variants that are found in allele count (AC) < 40 individuals for 245,388 short read WGS samples.
Extended Data Fig. 5 Ancestry specific HLA-DQB1 ( rs9273363 ) locus associations in 231,442 unrelated individuals.
Phenome-wide (PheWAS) associations highlight ancestry specific consequences across ancestries.
Extended Data Fig. 6 Ancestry specific TCF7L2 ( rs7903146 ) locus associations in 231,442 unrelated individuals.
Phenome-wide (PheWAS) associations highlight diabetic consequences across ancestries.
Supplementary information
Supplementary information.
Supplementary Figs. 1–7, Tables 1–8 and Note.
Reporting Summary
Supplementary dataset 1.
Associations of ACKR1, HLA-DQB1 and TCF7L2 loci with all Phecodes stratified by genetic ancestry.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/ .
Reprints and permissions
About this article
Cite this article.
The All of Us Research Program Genomics Investigators. Genomic data in the All of Us Research Program. Nature 627 , 340–346 (2024). https://doi.org/10.1038/s41586-023-06957-x
Download citation
Received : 22 July 2022
Accepted : 08 December 2023
Published : 19 February 2024
Issue Date : 14 March 2024
DOI : https://doi.org/10.1038/s41586-023-06957-x
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
‘all of us’ genetics chart stirs unease over controversial depiction of race.
Nature (2024)
Global genomic diversity for All of Us
Nature Reviews Genetics (2024)
By submitting a comment you agree to abide by our Terms and Community Guidelines . If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing: Translational Research newsletter — top stories in biotechnology, drug discovery and pharma.

National Institutes of Health U.S. Dept. of Health & Human Services
There's a gap in medical research that only you can fill.

The All of Us Research Program has a simple mission. We want to speed up health research breakthroughs. To do this, we’re asking one million people to share health information. In the future, researchers can use this to conduct thousands of health studies.
What Is the All of Us Research Program?
All of Us is a new research program from the National Institutes of Health (NIH). The goal is to advance precision medicine. Precision medicine is health care that is based on you as an individual. It takes into account factors like where you live, what you do, and your family health history. Precision medicine’s goal is to be able to tell people the best ways to stay healthy. If someone does get sick, precision medicine may help health care teams find the treatment that will work best.
To get there, we need one million or more people. Those who join will share information about their health over time. Researchers will study this data. What they learn could improve health for generations to come. Participants are our partners. We’ll share information back with them over time.
How It Works
Participants Share Data
Participants share health data online. This data includes health surveys and electronic health records. Participants also may be asked to share physical measurements and blood and urine samples.

Data Is Protected
Personal information, like your name, address, and other things that easily identify participants will be removed from all data. Samples—also without any names on them—are stored in a secure biobank.

Researchers Study Data
In the future, approved researchers will use this data to conduct studies. By finding patterns in the data, they may make the next big medical breakthroughs.

Participants Get Information
Participants will get information back about the data they provide, which may help them learn more about their health.

Researchers Share Discoveries
Research may help in many ways. It may help find the best ways for people to stay healthy. It may also help create better tests and find the treatments that will work best for different people.
What Will Participants Do?
When you join the All of Us Research Program, you will be asked to enroll, give consent, and agree to share health records. You can do this online, or at one of our partner centers.
If you take part, you will be asked to complete health surveys. You may be asked for physical measurements and biosamples (blood and urine samples).
You may be invited to share more data in the future, through additional health surveys, health trackers, or other research studies.

Precision Medicine Initiative, PMI, All of Us , the All of Us logo, and "The Future of Health Begins With You" are service marks of the U.S. Department of Health & Human Services. The All of Us platform is for research only and does not provide medical advice, diagnosis or treatment.

The All of Us Researchers Convention will be held April 3-4, 2024.
The free, virtual event is open to researchers across all disciplines and career stages. It provides an opportunity for researchers who use All of Us data to showcase their work for others who share their interests in precision medicine.

About the Researchers Convention
The All of Us Researchers Convention is organized by Pyxis Partners in collaboration with the All of Us Division of Engagement and Outreach. The division partners with community organizations nationwide to foster relationships with participants, researchers, and health care providers. The convention and other researcher engagement activities are central to building a diverse community of researchers.
This platform is supported by Pyxis Partners . The All of Us Researchers Convention is funded by the Division of Engagement and Outreach, All of Us Research Program, National Institutes of Health, under Award Number OD028404.
The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.

Description
The All of Us Research Program is a historic effort to gather information from one million or more people living in the United States. All of Us is a research program funded by the National Institutes of Health (NIH). The mission of the All of Us Research Program is to accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us.

- Age : 18 and up
- Gender : Any
- Keywords : Diversity, Personalized Medicine
- Type : Observational
- Target : 1,000,000 Participants

Investigator
Lucila Ohno-Machado, M.D., Ph.D., M.B.A. UC San Diego Health View Profile Co-Investigators Faculty Collaborators
What to expect if you participate
- What will we ask you to do? First, we will ask you to consent to participate in the study and to share your data with us. This is completed on the computer.
- If you decide to join All of Us, we will gather data about you. We will gather some of the data from you directly. We will gather some of the data from elsewhere.
- We will ask you to answer questions on the computer. We will ask you for data like your name, date of birth, and contact information. We will also ask questions about your health, family, home, and work.
- We will ask you to come to UC San Diego Health for a research visit so that we can measure you. Measurements include things like your height, weight, blood pressure, and heart rate. We will ask you to give a blood and urine sample after we take your physical measurements.
- We will ask you to allow us to access your health records.
- Will you ask me to do anything else? We will ask you to answer a few questions every year or so on the computer.
Benefits of participating
- You will not get direct medical benefit from taking part in All of Us.
- That said, you may indirectly benefit from taking part in All of Us. For example, we will provide ways for you to get access to all of the data you share with us and some of the results about you. This information may be interesting to you.
- You may learn about your health. You will be able to share your All of Us information with your healthcare provider if you choose. Finally, you will be helping researchers make discoveries that may help future generations.
Risks of participating
- The main risk of taking part in All of Us is to your privacy. A data breach is when someone sees or uses data without permission. If there is a data breach, someone could see or use the data we have about you.
- We will gather data from you through the All of Us app and/or website. You may be asked to wear a fitness tracker. There is a risk to your privacy whenever you use an app, website, or fitness tracker.
- Researchers will use basic facts like your race, ethnic group, and sex in their studies. This data helps researchers learn if the things that affect health are the same in different groups of people.
- If you give a blood sample, the most common risks are brief pain and bruising. Some people may become dizzy or feel faint. There is also a small risk of infection.
- Taking part in All of Us may have risks that we don't know about yet. We will tell you if we learn anything that might change your decision to take part.
Compensation
$25 gift card upon completion of your visit
Appointment length An in-person visit of about 30 minutes to 1 hour. Providing online updates to your health and lifestyle information from time to time. The entire research program is designed to last 10 years or more. Recruitment period From Aug. 23, 2017 to March 31, 2023
851 School of Medicine Building 2 (“The Hut”) 9500 Gilman Drive La Jolla, CA 92093 (Behind the Medical Education and Telemedicine Building) Walk-in Hours: Monday-Friday 8:00am-3:00pm By appointment: Monday-Friday 7:00am-3:00pm; Saturdays 9:00-3:30pm All Visitors Welcome Phone: 858-265-1711
UC San Diego Health – La Jolla Internal Medicine --> UC San Diego Health – University Pacific Center 8899 University Center Lane, Suite 400 San Diego, CA 92122 By appointment only: Monday, Wednesday, Thursday, Friday 8:30am-11:00am, 1:00am-3:00pm; Tuesdays 1:00pm-3:00pm UC San Diego Health Patients Only Phone: 858-287-0512 UC San Diego Health – La Jolla Perlman Medical Offices and/or Gastroenterology Department 9350 Campus Point Drive La Jolla, CA 92037 By appointment only: 7:30am-2:00pm UC San Diego Health Patients Only Phone: 858-287-0530 UC San Diego Health – Neurological Institute
4510 Executive Drive, Suite 325 San Diego, CA 92121 Hours: Fridays only 8:00am-11:00am Neurological Institute Patients Only Phone: 858-265-1711
UC San Diego Health – Fourth & Lewis Medical Offices, Hillcrest 330 Lewis Street San Diego, CA 92103 Hours: Mondays only 8:00am-11:00am UC San Diego Health Patients Only Phone: 858-287-0507 UC San Diego Health – Hillcrest North – Gastroenterology Department 200 West Arbor Drive San Diego, CA 92103 By appointment only: Monday-Thursday 7:00am-1:00pm; Fridays 7:00am-11:00am All Visitors Welcome Phone: 858-287-0507 El Centro Regional Medical Center - Outpatient Clinic 385 W. Main Street El Centro, CA 92243 Walk-in Hours: Monday-Friday 8:00am-3:00pm Email: [email protected] Phone: 760-490-1215 San Diego Blood Bank Locations If it is more convenient for you, we can assist you with scheduling your visit at one of our San Diego Blood Bank locations. Gateway Donor Center
3636 Gateway Center Avenue San Diego, CA 92102 Hours: Monday-Friday 8:00am-2:30pm Phone: 833-271-1388
Sabre Springs Donor Center 12640 Sabre Springs Parkway San Diego, CA 92128 Hours: Tuesday-Thursday 10:30am-1:00pm; Friday 8:30am-1:00pm Phone: 833-271-1388 East County Donor Center
776 Arnele Avenue El Cajon, CA 92020 Hours: Monday and Friday 8:30am-1:00pm, Tuesday and Thursday 10:30am-1:00pm, Wednesday 10:00am-1:00pm Phone: 833-271-1388
North County Donor Center
338 West El Norte Parkway, Suite J Escondido, CA 92026 Hours: Wednesday 10:00am-1:00pm, Thursday 10:30am-1:00pm, Friday 8:30am-1:00pm Phone: 833-271-1388

© Copyright UC San Diego Health 2019
Precision Medicine Initiative, PMI, All of Us , the All of Us logo, and “The Future of Health Begins with You” are service marks of the U.S. Department of Health and Human Services.
© Copyright UC San Diego Health 2019
- Skip to primary navigation
- Skip to main content
Research that reaches everyone
Students from all disciplines gain opportunities with new undergraduate student research program.
April 16, 2024
White coats, labs, microscopes, chemical reactions and science are typically associated with the word “research.” But research occurs in all 15 departments within the Texas A&M University College of Agriculture and Life Sciences . From economics, ethical leadership, livestock, communications and water quality, research permeates every aspect of agriculture and life sciences.
“Research isn’t confined to STEM-related fields,” said Craig Coates, Ph.D., associate dean for programmatic success for the College. “Regardless of your major, there are opportunities for undergraduate research.”
This all-encompassing approach to research is precisely what Coates advocates through the Undergraduate Student Research Program . The program is tailored to support students interested in conducting research across any discipline within the College.
Curiosity is key
To foster and honor excellence in research, Coates and key College leadership devised the Undergraduate Student Research Program to support up to 30 research scholars each year. Each successful scholar will be awarded a $1,000 scholarship, while faculty mentoring these students will receive up to $500 to cover expenses such as materials, travel and other related costs.

To qualify for the program, undergraduate students within the College must be either current sophomores or juniors with an expected graduation date between May 2025 and May 2026. Students must also register for a research course with their faculty mentor, complete the online Responsible Conduct of Research training and submit a research proposal approved by their research faculty mentor by April 30.
Selected scholars will engage in research alongside their faculty mentor during both the fall and spring semesters. They will then have the opportunity to present their research at a symposium in the spring.
“Embarking on a research program might seem daunting,” Coates said. “However, possessing curiosity is crucial for success, particularly in research. My advice for students is to explore the online profiles of faculty members within their department to discover areas of research that pique their interest. What sparks your curiosity? Your research journey can be ignited by a subject you’re passionate about.”
To learn more about the Undergraduate Student Research Program and to apply, visit the program’s website .
- Resources for Press & Media
- Story Suggestion

Apr. 15, 2024
New rice research explores why we remember what we remember.

We’ve all been in a similar situation — you lock your front door for the umpteenth time in a given week only to panic minutes later when you’re driving to work as you struggle to remember if you actually locked the door. If this sounds familiar, you’re not alone, and you’re also not losing your mind. A new study published in Neurobiology of Learning and Memory by Rice University psychologists found that certain experiences are better remembered by most people, while other experiences, like locking the door behind us, are more easily forgotten.

However, the story isn’t quite that simple, according to researchers Fernanda Morales-Calva , a Rice graduate student, and Stephanie Leal , assistant professor of psychological sciences. They conducted the study to better understand just how human memory works. They said humans tend to focus on remembering certain aspects within an experience more than others such as the big picture of what happened rather than the details. “Struggling to remember is one of those things we all experience,” Morales-Calva said. “But when it comes to understanding memory, there’s a lot to be discovered about how it actually works. And there’s a new area of memory research that’s trying to tap into why we remember certain things better than others.” For example, Morales-Calva said people looking back on the last year may recall doing a lot of different things, but only a few of them might really stand out in great detail. “Previous research has found that these memorable experiences for one person are very likely memorable for another person, like birthday parties, deaths of a loved one and more,” Leal said. “These are often positive or negative experiences. This knowledge has helped us design research studies looking at memory performance.” The researchers evaluated memory by showing pictures to their study participants. During a memory test, some of these images were repeated, some were brand new, while others were very similar and difficult to distinguish from one another. These similar images were meant to interfere with memory, kind of like the similar daily experiences such as trying to remember if the door is locked. Memorable images were identified as the ones participants were most likely to recall. Morales-Calva and Leal found that while participants correctly remembered the most memorable images, this effect was lost after 24 hours. This was especially true when remembering positive experiences, suggesting these experiences are memorable at first but more prone to be forgotten. “While we feel like we know what types of experiences are memorable, we really don’t know what features of a memory are remembered best in the long term,” Morales-Calva said. “We often think emotional memories are better remembered, but in fact gist versus detail trade-offs exist where the central features of the memory are enhanced while details may be forgotten.” So if you’re one of the many people in the world who can’t remember if five minutes ago you put down your garage door or swallowed your medicine, the researchers said you’re not alone. “Our brains can’t possibly remember everything we experience, and so we have to do a bit of selective forgetting for information that isn’t as important,” Leal said. “This study helps us get closer to understanding why we remember what we remember.” Morales-Calva and Leal said they hope their findings will offer new insights about how memory works, why some things are memorable and others are not. They hope future studies will consider the complexity of memory in everyday life, including consideration of the emotional content, time that has passed since the experience and perceptual features of memory that may have significant impacts on what we remember. The article, “Emotional modulation of memorability in mnemonic discrimination,” is available online .
- Share full article

Chinese Company Under Congressional Scrutiny Makes Key U.S. Drugs
Lawmakers raising national security concerns and seeking to disconnect a major Chinese firm from U.S. pharmaceutical interests have rattled the biotech industry. The firm is deeply involved in development and manufacturing of crucial therapies for cancer, cystic fibrosis, H.I.V. and other illnesses.
A WuXi Biologics facility in Wuxi, China. WuXi AppTec and an affiliated company, WuXi Biologics, have received millions of dollars in tax incentives to build sprawling research and manufacturing sites in Massachusetts and Delaware. Credit... Imaginechina Limited, via Alamy
Supported by

By Christina Jewett
- April 15, 2024
A Chinese company targeted by members of Congress over potential ties to the Chinese government makes blockbuster drugs for the American market that have been hailed as advances in the treatment of cancers, obesity and debilitating illnesses like cystic fibrosis.
WuXi AppTec is one of several companies that lawmakers have identified as potential threats to the security of individual Americans’ genetic information and U.S. intellectual property. A Senate committee approved a bill in March that aides say is intended to push U.S. companies away from doing business with them.
But lawmakers discussing the bill in the Senate and the House have said almost nothing in hearings about the vast scope of work that WuXi does for the U.S. biotech and pharmaceutical industries — and patients. A New York Times review of hundreds of pages of records worldwide shows that WuXi is heavily embedded in the U.S. medicine chest, making some or all of the main ingredients for multibillion-dollar therapies that are highly sought to treat cancers like some types of leukemia and lymphoma as well as obesity and H.I.V.
The Congressional spotlight on the company has rattled the pharmaceutical industry, which is already struggling with widespread drug shortages now at a 20-year high . Some biotech executives have pushed back, trying to impress on Congress that a sudden decoupling could take some drugs out of the pipeline for years.
WuXi AppTec and an affiliated company, WuXi Biologics grew rapidly, offering services to major U.S. drugmakers that were seeking to shed costs and had shifted most manufacturing overseas in the last several decades.
WuXi companies developed a reputation for low-cost and reliable work by thousands of chemists who could create new molecules and operate complex equipment to make them in bulk. By one estimate, WuXi has been involved in developing one-fourth of the drugs used in the United States. WuXi AppTec reported earning about $3.6 billion in revenue for its U.S. work.
“They have become a one-stop shop to a biotech,” said Kevin Lustig, founder of Scientist.com, a clearinghouse that matches drug companies seeking research help with contractors like WuXi.
WuXi AppTec and WuXi Biologics have also received millions of dollars in tax incentives to build sprawling research and manufacturing sites in Massachusetts and Delaware that local government officials have welcomed as job and revenue generators. One WuXi site in Philadelphia was working alongside a U.S. biotech firm to give patients a cutting-edge therapy that would turbocharge their immune cells to treat advanced skin cancers.
The tension has grown since February, when four lawmakers asked the Commerce, Defense and Treasury Departments to investigate WuXi AppTec and affiliated companies, calling WuXi a “giant that threatens U.S. intellectual property and national security.”
A House bill called the Biosecure Act linked the company to the People’s Liberation Army, the military arm of the Chinese Communist Party. The bill claims WuXi AppTec sponsored military-civil events and received military-civil fusion funding.
Richard Connell, the chief operating officer of WuXi AppTec in the United States and Europe, said the company participates in community events, which do not “imply any association with or endorsement of a government institution, political party or policy such as military-civil fusion.” He also said shareholders do not have control over the company or access to nonpublic information.

Last month, after a classified briefing with intelligence staff, the Senate homeland security committee advanced a bill by a vote of 11 to 1: It would bar companies from receiving government contracts for work with Wuxi, but would allow the companies to still obtain contracts for unrelated projects. Government contracts with drugmakers are generally limited, though they were worth billions of dollars in revenue to companies that responded to the Covid-19 pandemic.
Mr. Connell defended the company’s record, saying the proposed legislation “relies on misleading allegations and inaccurate assertions against our company.”
WuXi operates in a highly regulated environment by “multiple U.S. federal agencies — none of which has placed our company on any sanctions list or designated it as posing a national security risk,” Mr. Connell said. WuXi Biologics did not respond to requests for comment.
Smaller biotech companies, which tend to rely on government grants and have fewer reserves, are among the most alarmed. Dr. Jonathan Kil, the chief executive of Seattle-based Sound Pharmaceuticals, said WuXi has worked alongside the company for 16 years to develop a treatment for hearing loss and tinnitus, or ringing in the ear. Finding another contractor to make the drug could set the company back two years, he said.
“What I don’t want to see is that we get very anti-Chinese to the point where we’re not thinking correctly,” Dr. Kil said.
It is unclear whether a bill targeting WuXi will advance at all this year. The Senate version has been amended to protect existing contracts and limit supply disruptions. Still, the scrutiny has prompted some drug and biotechnology companies to begin making backup plans.
Peter Kolchinsky, managing partner of RA Capital Management, estimated that half of the 200 biotech companies in his firm’s investment portfolio work with WuXi.
“Everyone is likely considering moving away from Wuxi and China more broadly,” he said in an email. “Even though the current versions of the bill don’t create that imperative clearly, no one wants to be caught flat-footed in China if the pullback from China accelerates.”
The chill toward China extends beyond drugmakers. U.S. companies are receiving billions of dollars in funding under the CHIPS Act, a federal law aimed at bringing semiconductor manufacturing stateside.
For the last several years, U.S. intelligence agencies have been warning about Chinese biotech companies in general and WuXi in particular. The National Counterintelligence and Security Center, the arm of the intelligence community charged with warning companies about national security issues, raised alarms about WuXi’s acquisition of NextCODE, an American genomic data company.
Though WuXi later spun off that company, a U.S. official said the government remains skeptical of WuXi’s corporate structure, noting that some independent entities have overlapping management and that there were other signs of the Chinese government’s continuing control or influence over WuXi.
Aides from the Senate homeland security committee said their core concerns are about the misuse of Americans’ genomic data, an issue that’s been more closely tied to other companies named in the bill.
Aides said the effort to discourage companies from working with WuXi and others was influenced by the U.S. government’s experience with Huawei, a Chinese telecommunications giant. By the time Congress acted on concerns about Huawei’s access to Americans’ private information, taxpayers had to pay billions of dollars to tear Huawei’s telecommunication equipment out of the ground.
Yet WuXi has far deeper involvement in American health care than has been discussed in Congress. Supply chain analytics firms QYOBO and Pharm3r, and some public records, show that WuXi and its affiliates have made the active ingredients for critical drugs.
They include Imbruvica, a leukemia treatment sold by Janssen Biotech and AbbVie that brought in $5.9 billion in worldwide revenue in 2023. WuXi subsidiary factories in Shanghai and Changzhou were listed in government records as makers of the drug’s core ingredient, ibrutinib.
Dr. Mikkael A. Sekeres, chief of hematology at the University of Miami Health System, called that treatment for chronic lymphocytic leukemia “truly revolutionary” for replacing highly toxic drugs and extending patients’ lives.
Janssen Biotech and AbbVie, partners in selling the drug, declined to comment.
WuXi Biologics also manufactures Jemperli, a GSK treatment approved by the Food and Drug Administration last year for some endometrial cancers. In combination with standard therapies, the drug improves survival in patients with advanced disease, said Dr. Amanda Nickles Fader, president of the Society of Gynecologic Oncology.
“This is particularly important because while most cancers are plateauing or decreasing in incidence and mortality, endometrial cancer is one of the only cancers globally” increasing in both, Dr. Fader said.
GSK declined to comment.
The drug that possibly captures WuXi’s most significant impact is Trikafta, manufactured by an affiliate in Shanghai and Changzhou to treat cystic fibrosis, a deadly disease that clogs the lungs with debilitating, thick mucus. The treatment is credited with clearing the lungs and extending by decades the life expectancy of about 40,000 U.S. residents. It also had manufacturers in Italy, Portugal and Spain.
The treatment has been so effective that the Make-A-Wish Foundation stopped uniformly granting wishes to children with cystic fibrosis. Trikafta costs about $320,000 a year per patient and has been a boon for Boston-based Vertex Pharmaceuticals and its shareholders, with worldwide revenue rising to $8.9 billion last year from $5.7 billion in 2021, according to a securities filing .
Trikafta “completely transformed cystic fibrosis and did it very quickly,” said Dr. Meghan McGarry, a University of California San Francisco pulmonologist who treats children with the condition. “People came off oxygen and from being hospitalized all the time to not being hospitalized and being able to get a job, go to school and start a family.”
Vertex declined to comment.
Two industry sources said WuXi plays a role in making Eli Lilly’s popular obesity drugs. Eli Lilly did not respond to requests for comment. WuXi companies also make an infusion for treatment-resistant H.I.V., a drug for advanced ovarian cancer and a therapy for adults with a rare disorder called Pompe disease.
WuXi is known for helping biotech firms from the idea stage to mass production, Dr. Kolchinsky said. For example, a start-up could hypothesize that a molecule that sticks to a certain protein might cure a disease. The company would then hire WuXi chemists to create or find the molecule and test it in petri dishes and animals to see whether the idea works — and whether it’s safe enough for humans.
“Your U.S. company has the idea and raises the money and owns the rights to the drug,” Dr. Kolchinsky said. “But they may count on WuXi or similar contractors for almost every step of the process.”
WuXi operates large bioreactors and manufactures complex peptide, immunotherapy and antibody drugs at sprawling plants in China.
WuXi AppTec said it has about 1,900 U.S. employees. Officials in Delaware gave the company $19 million in tax funds in 2021 to build a research and drug manufacturing site that is expected to employ about 1,000 people when fully operational next year, public records and company reports show.
Mayor Kenneth L. Branner Jr. of Middletown, Del., called it “one of those once-in-a-lifetime opportunities to land a company like this,” according to a news report when the deal was approved.
In 2022, the lieutenant governor of Massachusetts expressed a similar sentiment when workers placed the final steel beam on a WuXi Biologics research and manufacturing plant in Worcester. Government officials had approved roughly $11.5 million in tax breaks to support the project. The company announced this year that it would double the site’s planned manufacturing capacity in response to customer demand.
And in Philadelphia, a WuXi Advanced Therapies site next to Iovance Biotherapeutics was approved by regulators to help process individualized cell therapies for skin cancer patients. Iovance has said it is capable of meeting demand for the therapies independently.
By revenue, WuXi Biologics is one of the top five drug development and manufacturing companies worldwide, according to Statista , a data analytics company. A WuXi AppTec annual report showed that two-thirds of its revenue came from U.S. work.
Stepping away from WuXi could cause a “substantial slowdown” in drug development for a majority of the 105 biotech companies surveyed by BioCentury , a trade publication. Just over half said it would be “extremely difficult” to replace China-based drug manufacturers.
BIO, a trade group for the biotechnology industry, is also surveying its members about the impact of disconnecting from WuXi companies. John F. Crowley, BIO’s president, said the effects would be most difficult for companies that rely on WuXi to manufacture complex drugs at commercial scale. Moving such an operation could take five to seven years.
“We have to be very thoughtful about this so that we first do no harm to patients,” Mr. Crowley said. “And that we don’t slow or unnecessarily interfere with the advancement of biomedical research.”
Julian E. Barnes contributed reporting, and Susan C. Beachy contributed research.
Christina Jewett covers the Food and Drug Administration, which means keeping a close eye on drugs, medical devices, food safety and tobacco policy. More about Christina Jewett
Advertisement
- Skip to main content
- Keyboard shortcuts for audio player

- LISTEN & FOLLOW
- Apple Podcasts
- Google Podcasts
- Amazon Music
- Amazon Alexa
Your support helps make our show possible and unlocks access to our sponsor-free feed.
The order your siblings were born in may play a role in identity and sexuality

Selena Simmons-Duffin
Rachel Carlson

Rebecca Ramirez

It's National Siblings Day ! To mark the occasion, guest host Selena Simmons-Duffin is exploring a detail very personal to her: How the number of older brothers a person has can influence their sexuality.
Scientific research on sexuality has a dark history, with long-lasting harmful effects on queer communities. Much of the early research has also been debunked over time. But not this "fraternal birth order effect." The fact that a person's likelihood of being gay increases with each older brother has been found all over the world – from Turkey to North America, Brazil, the Netherlands and beyond. Today, Selena gets into all the details: What this effect is, how it's been studied and what it can (and can't) explain about sexuality.
Interested in the science of our closest relatives? Check out more stories in NPR's series on the Science of Siblings .
Email us at [email protected] — we'd love to hear from you.
Listen to Short Wave on Spotify , Apple Podcasts and Google Podcasts .
Listen to every episode of Short Wave sponsor-free and support our work at NPR by signing up for Short Wave+ at plus.npr.org/shortwave .
This episode was produced by Rachel Carlson. It was edited by Rebecca Ramirez and fact-checked by Brit Hanson. Maggie Luthar was the audio engineer.
More from the Science of Siblings series:
- The origin story of National Sibling Day is a celebration of love — and grief
- In the womb, a brother's hormones can shape a sister's future
- These identical twins both grew up with autism, but took very different paths
- birth order

IMAGES
VIDEO
COMMENTS
The National Institutes of Health's All of Us Research Program is building one of the largest biomedical data resources of its kind. The All of Us Research Hub stores health data from a diverse group of participants from across the United States.. Register for the Researcher Workbench to access data and tools to conduct health research and improve understanding of health and disease.
The All of Us Research Program is a historic effort to gather data from one million or more people living in the United States to accelerate research and improve health. By taking into account individual differences in genes, environment, and lifestyle, researchers will uncover paths toward delivering precision medicine.
About. The All of Us Research Program is a historic effort to collect and study data from one million or more people living in the United States. The goal of the program is better health for all of us. Our mission is to accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us.
What All of Uscan do for you. If you join All of Us and provide biosamples, like blood or saliva, you may choose to learn more about your DNA: Your genetic ancestry. Your risk for certain hereditary diseases. Your body's reaction to certain medicines. There is no cost to participate other than some of your time.
Also, all dates are systematically shifted backwards by a random number between 1 and 365, and data from participants over the age of 89 are removed. The All of Us Research Program data will be accessed for research strictly using the Researcher Workbench (researchallofus.org). External data can be brought into this secure environment; however ...
The group concluded its work in September 2015 with a detailed report. The report provided a framework for setting up the All of Us Research Program. Precision medicine: Is based on you as an individual. Takes into account your environment (where you live), lifestyle (what you do), and your family health history and genetic makeup.
All of Us is a Research Program of the National Institutes of Health (NIH). The mission of All of Us is to accelerate health research and medical breakthroughs, enabling individualized prevention, treatment, and care for all of us by building dataset of one million or more volunteers nationwide who will sign up to share their health information over time.
Precision Medicine Initiative, PMI, All of Us, the All of Us logo, and "The Future of Health Begins With You" are service marks of the U.S. Department of Health and Human Services.The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
Precision Medicine Initiative, PMI, All of Us, the All of Us logo, and "The Future of Health Begins With You" are service marks of the U.S. Department of Health and Human Services. The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
All of Us is a new kind of research program. We're building the largest and most inclusive health research initiative of its kind. We're collecting health data from over a million people in an effort to help researchers learn more about health. We work with communities that have not been represented in the past to understand health ...
All of Us, a research program from the National Institutes of Health (NIH), aims to have one million or more volunteers who reflect the diversity of the United States. Researchers will use the participant data collected to study many different conditions, including cancer, diabetes, depression, asthma, and other diseases.
The All of Us Research Program and its many participant partners are leading the way toward more equitable representation in medical research. About half of this new genomic information comes from people who self-identify with a racial or ethnic minority group. That's extremely important because, until now, over 90 percent of participants in ...
Precision Medicine Initiative, PMI, All of Us, the All of Us logo, and "The Future of Health Begins With You" are service marks of the U.S. Department of Health and Human Services.The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
About the All of Us Research Program. The National Institutes of Health's All of Us Research Program is a historic effort to collect and study data from at least one million people living in the United States. The goal of All of Us is to speed up health research discoveries, enabling new kinds of individualized health care. To make this possible, the program is building one of the world's ...
To accelerate health research, All of Us is committed to curating and releasing research data early and often 6.Less than five years after national enrolment began in 2018, this fifth data release ...
Diversity. The program is enrolling a large group of people that reflects the diversity of the United States. This includes people who haven't taken part in or have been left out of health research before. All of Us welcomes participants of all backgrounds and walks of life, from all regions of the country, whether they are healthy or sick ...
The All of Us Research Program is a large research program from the National Institutes of Health. The goal is to help researchers understand more about why people get sick or stay healthy. People who join will share information. This might be about their health, habits, and what it's like where they live.
The All of Us Researchers Convention is funded by the Division of Engagement and Outreach, All of Us Research Program, National Institutes of Health, under Award Number OD028404. The All of Us platform is for research only and does not provide medical advice, diagnosis, or treatment.
United States House Committee on Oversight and Accountability. WASHINGTON—The Select Subcommittee on the Coronavirus Pandemic held a hearing on "Investigating the Origins of COVID-19" to gather facts about the origination of the virus that has claimed nearly seven million lives globally. At the hearing, several of the witnesses pointed to how the science, facts, and evidence point to a ...
All of Us - UC San Diego Onsite Clinic. 851 School of Medicine Building 2 ("The Hut") 9500 Gilman Drive. La Jolla, CA 92093. (Behind the Medical Education and Telemedicine Building) Walk-in Hours: Monday-Friday 8:00am-3:00pm. By appointment: Monday-Friday 7:00am-3:00pm; Saturdays 9:00-3:30pm. All Visitors Welcome.
Students from all disciplines gain opportunities with new Undergraduate Student Research Program. April 16, 2024. White coats, labs, microscopes, chemical reactions and science are typically associated with the word "research.". But research occurs in all 15 departments within the Texas A&M University College of Agriculture and Life Sciences.
More than 1,195,000 people have registered with the program by creating online accounts at JoinAllofUs.org, beginning the enrollment process. The snapshots below highlight participants in the All of Us Research Program. The following numbers are approximated to protect participants' privacy. Numbers reflect data collected through April 15, 2024.
"Previous research has found that these memorable experiences for one person are very likely memorable for another person, like birthday parties, deaths of a loved one and more," Leal said. "These are often positive or negative experiences. This knowledge has helped us design research studies looking at memory performance."
June 23, 2022 — A new report shows more than 18 million Americans (8.3 million males and 9.7 million females) with a history of cancer were living in the United States as of January 1, 2022 ...
The 2024 nationwide tour, called "the All of Us Journey," begins April 16th with stops planned in California to the Midwest and throughout New England. The All of Us Journey mobile exhibit shares information about the program's goal to build one of the most diverse health databases in history. With robust data, researchers can learn more ...
The goal of All of Us is to speed up health research discoveries, enabling new kinds of individualized health care.To make this possible, the program is building one of the world's largest and most diverse databases for health research. By working with participants across the country, collecting many types of information over time, and building a data platform that many researchers can use ...
A New York Times review of hundreds of pages of records worldwide shows that WuXi is heavily embedded in the U.S. medicine chest, making some or all of the main ingredients for multibillion-dollar ...
Scientific research on sexuality has a dark history, with long-lasting harmful effects on queer communities. Much of the early research has also been debunked over time. But not this "fraternal ...
The All of Us Research Program Genomics Investigators. Nature. 2024 Feb 19. doi: 10.1038/s41586-023-06957-x. Browse or Search all Publications. Sort By: Date. Title. Mental Health Conditions Associated With Strabismus in a Diverse Cohort of US Adults.
Unlike many research studies that focus on a specific disease or population, the All of Us Research Program will provide a national research resource to inform thousands of research questions, covering a wide variety of health conditions. A diverse cohort of 1 million or more participants will contribute data from electronic health records (EHRs), biospecimens, surveys, and other measures to ...