
Statistics Made Easy

How to Fix in R: missing values are not allowed in subscripted assignments
One error message you may encounter when using R is:
This error usually occurs when you attempt to assign values in one column using values from another column, but there happen to be NA values present.
The following example shows how to resolve this error in practice.
How to Reproduce the Error
Suppose we create the following data frame in R:
Now suppose we attempt to assign a value of 10 to each row in column B where the corresponding value in column A is equal to 5:
We receive an error because there are NA values in column A and we’re explicitly told in the error message that missing values are not allowed in subscripted assignments of data frames .
How to Avoid the Error
There are two ways to avoid this error.
1. Use %in% Operator
One way to avoid this error is to use the %in% operator when performing the assignment:
Notice that a value of 10 has been assigned to each row in column B where the corresponding value in column A is equal to 5 and we don’t receive any error.
2. Use is.na()
Another way to avoid this error is to use the is.na() function when performing the assignment:
Once again we’re able to assign a value of 10 to each row in column B where the corresponding value in column A is equal to 5 and we don’t receive any error.
Additional Resources
The following tutorials explain how to fix other common errors in R:
How to Fix in R: Arguments imply differing number of rows How to Fix in R: error in select unused arguments How to Fix in R: replacement has length zero
Published by Zach
Leave a reply cancel reply.
Your email address will not be published. Required fields are marked *

How to Solve R Error: missing values are not allowed in subscripted assignments of data frames
by Suf | Programming , R , Tips
This error occurs when you attempt to subscript a data frame that contains NA values. You can solve this error by excluding the NA values during the subscript operation, for example:
This tutorial will go through the error in detail and how to solve it with code examples.
Table of contents
Consider the following 100 row data frame:
The first variable contains an NA value and samples from the normal distribution. The second variable has five integer values that serve as categories. Let’s look at the head of the data frame:
Next, we will try to replace the value in the x column that are less than zero with zero.
Let’s run the code to see the result:
The error occurs because the x column contains NA values.
We can solve this error by using the is.na function in the subscript operation.
By using !is.na() we are excluding the NA values during the subscript operation. Let’s run the code to see the result:
We successfully substituted the values less than zero with zero in the x column.
Congratulations on reading to the end of this tutorial!
For further reading on R related errors, go to the articles:
- How to Solve R Error in file(file, “rt”) cannot open the connection
- How to Solve R Error: aesthetics must be either length 1 or the same as the data
- How to Solve R Error: continuous value supplied to discrete scale
Go to the online courses page on R to learn more about coding in R for data science and machine learning.
Have fun and happy researching!
Share this:
- Click to share on Facebook (Opens in new window)
- Click to share on LinkedIn (Opens in new window)
- Click to share on Reddit (Opens in new window)
- Click to share on Pinterest (Opens in new window)
- Click to share on Telegram (Opens in new window)
- Click to share on WhatsApp (Opens in new window)
- Click to share on Twitter (Opens in new window)
- Click to share on Tumblr (Opens in new window)
How to Fix in R: missing values are not allowed in subscripted assignments
One error message you may encounter when using R is:
This error usually occurs when you attempt to assign values in one column using values from another column, but there happen to be NA values present.
The following example shows how to resolve this error in practice.
How to Reproduce the Error
Suppose we create the following data frame in R:
Now suppose we attempt to assign a value of 10 to each row in column B where the corresponding value in column A is equal to 5:
We receive an error because there are NA values in column A and we’re explicitly told in the error message that missing values are not allowed in subscripted assignments of data frames .
How to Avoid the Error
There are two ways to avoid this error.
1. Use %in% Operator
One way to avoid this error is to use the %in% operator when performing the assignment:
Notice that a value of 10 has been assigned to each row in column B where the corresponding value in column A is equal to 5 and we don’t receive any error.
2. Use is.na()
Another way to avoid this error is to use the is.na() function when performing the assignment:
Once again we’re able to assign a value of 10 to each row in column B where the corresponding value in column A is equal to 5 and we don’t receive any error.
Additional Resources
The following tutorials explain how to fix other common errors in R:
How to Fix in R: Arguments imply differing number of rows How to Fix in R: error in select unused arguments How to Fix in R: replacement has length zero
How to Find the Median of a Box Plot (With Examples)
How to create a list of lists in r (with example), related posts, how to create a stem-and-leaf plot in spss, how to create a correlation matrix in spss, how to convert date of birth to age..., excel: how to highlight entire row based on..., how to add target line to graph in..., excel: how to use if function with negative..., excel: how to use if function with text..., excel: how to use greater than or equal..., excel: how to use if function with multiple..., pandas: how to rename only the last column....
- Introduction
- Foundations
- Data structures
- OO field guide
- Environments
- Exceptions and debugging
- Functional programming
- Functionals
- Function operators
- Metaprogramming
- Non-standard evaluation
- Expressions
- Domain specific languages
- Performant code
- Performance
- R's C interface
Advanced R by Hadley Wickham
R’s subsetting operators are powerful and fast. Mastery of subsetting allows you to succinctly express complex operations in a way that few other languages can match. Subsetting is hard to learn because you need to master a number of interrelated concepts:
The three subsetting operators.
The six types of subsetting.
Important differences in behaviour for different objects (e.g., vectors, lists, factors, matrices, and data frames).
The use of subsetting in conjunction with assignment.
This chapter helps you master subsetting by starting with the simplest type of subsetting: subsetting an atomic vector with [ . It then gradually extends your knowledge, first to more complicated data types (like arrays and lists), and then to the other subsetting operators, [[ and $ . You’ll then learn how subsetting and assignment can be combined to modify parts of an object, and, finally, you’ll see a large number of useful applications.
Subsetting is a natural complement to str() . str() shows you the structure of any object, and subsetting allows you to pull out the pieces that you’re interested in.
Take this short quiz to determine if you need to read this chapter. If the answers quickly come to mind, you can comfortably skip this chapter. Check your answers in answers .
What is the result of subsetting a vector with positive integers, negative integers, a logical vector, or a character vector?
What’s the difference between [ , [[ , and $ when applied to a list?
When should you use drop = FALSE ?
If x is a matrix, what does x[] <- 0 do? How is it different to x <- 0 ?
How can you use a named vector to relabel categorical variables?
Data types starts by teaching you about [ . You’ll start by learning the six types of data that you can use to subset atomic vectors. You’ll then learn how those six data types act when used to subset lists, matrices, data frames, and S3 objects.
Subsetting operators expands your knowledge of subsetting operators to include [[ and $ , focussing on the important principles of simplifying vs. preserving.
In Subsetting and assignment you’ll learn the art of subassignment, combining subsetting and assignment to modify parts of an object.
Applications leads you through eight important, but not obvious, applications of subsetting to solve problems that you often encounter in a data analysis.
It’s easiest to learn how subsetting works for atomic vectors, and then how it generalises to higher dimensions and other more complicated objects. We’ll start with [ , the most commonly used operator. Subsetting operators will cover [[ and $ , the two other main subsetting operators.
Atomic vectors
Let’s explore the different types of subsetting with a simple vector, x .
Note that the number after the decimal point gives the original position in the vector.
There are five things that you can use to subset a vector:
Positive integers return elements at the specified positions:
Negative integers omit elements at the specified positions:
You can’t mix positive and negative integers in a single subset:
Logical vectors select elements where the corresponding logical value is TRUE . This is probably the most useful type of subsetting because you write the expression that creates the logical vector:
If the logical vector is shorter than the vector being subsetted, it will be recycled to be the same length.
A missing value in the index always yields a missing value in the output:
Nothing returns the original vector. This is not useful for vectors but is very useful for matrices, data frames, and arrays. It can also be useful in conjunction with assignment.
Zero returns a zero-length vector. This is not something you usually do on purpose, but it can be helpful for generating test data.
If the vector is named, you can also use:
Character vectors to return elements with matching names.
Subsetting a list works in the same way as subsetting an atomic vector. Using [ will always return a list; [[ and $ , as described below, let you pull out the components of the list.
Matrices and arrays
You can subset higher-dimensional structures in three ways:
- With multiple vectors.
- With a single vector.
- With a matrix.
The most common way of subsetting matrices (2d) and arrays (>2d) is a simple generalisation of 1d subsetting: you supply a 1d index for each dimension, separated by a comma. Blank subsetting is now useful because it lets you keep all rows or all columns.
By default, [ will simplify the results to the lowest possible dimensionality. See simplifying vs. preserving to learn how to avoid this.
Because matrices and arrays are implemented as vectors with special attributes, you can subset them with a single vector. In that case, they will behave like a vector. Arrays in R are stored in column-major order:
You can also subset higher-dimensional data structures with an integer matrix (or, if named, a character matrix). Each row in the matrix specifies the location of one value, where each column corresponds to a dimension in the array being subsetted. This means that you use a 2 column matrix to subset a matrix, a 3 column matrix to subset a 3d array, and so on. The result is a vector of values:
Data frames
Data frames possess the characteristics of both lists and matrices: if you subset with a single vector, they behave like lists; if you subset with two vectors, they behave like matrices.
S3 objects are made up of atomic vectors, arrays, and lists, so you can always pull apart an S3 object using the techniques described above and the knowledge you gain from str() .
There are also two additional subsetting operators that are needed for S4 objects: @ (equivalent to $ ), and slot() (equivalent to [[ ). @ is more restrictive than $ in that it will return an error if the slot does not exist. These are described in more detail in the OO field guide .
Fix each of the following common data frame subsetting errors:
Why does x <- 1:5; x[NA] yield five missing values? (Hint: why is it different from x[NA_real_] ?)
What does upper.tri() return? How does subsetting a matrix with it work? Do we need any additional subsetting rules to describe its behaviour?
Why does mtcars[1:20] return an error? How does it differ from the similar mtcars[1:20, ] ?
Implement your own function that extracts the diagonal entries from a matrix (it should behave like diag(x) where x is a matrix).
What does df[is.na(df)] <- 0 do? How does it work?
Subsetting operators
There are two other subsetting operators: [[ and $ . [[ is similar to [ , except it can only return a single value and it allows you to pull pieces out of a list. $ is a useful shorthand for [[ combined with character subsetting.
You need [[ when working with lists. This is because when [ is applied to a list it always returns a list: it never gives you the contents of the list. To get the contents, you need [[ :
“If list x is a train carrying objects, then x[[5]] is the object in car 5; x[4:6] is a train of cars 4-6.” — @RLangTip
Because it can return only a single value, you must use [[ with either a single positive integer or a string:
Because data frames are lists of columns, you can use [[ to extract a column from data frames: mtcars[[1]] , mtcars[["cyl"]] .
S3 and S4 objects can override the standard behaviour of [ and [[ so they behave differently for different types of objects. The key difference is usually how you select between simplifying or preserving behaviours, and what the default is.
Simplifying vs. preserving subsetting
It’s important to understand the distinction between simplifying and preserving subsetting. Simplifying subsets returns the simplest possible data structure that can represent the output, and is useful interactively because it usually gives you what you want. Preserving subsetting keeps the structure of the output the same as the input, and is generally better for programming because the result will always be the same type. Omitting drop = FALSE when subsetting matrices and data frames is one of the most common sources of programming errors. (It will work for your test cases, but then someone will pass in a single column data frame and it will fail in an unexpected and unclear way.)
Unfortunately, how you switch between simplifying and preserving differs for different data types, as summarised in the table below.
Preserving is the same for all data types: you get the same type of output as input. Simplifying behaviour varies slightly between different data types, as described below:
Atomic vector : removes names.
List : return the object inside the list, not a single element list.
Factor : drops any unused levels.
Matrix or array : if any of the dimensions has length 1, drops that dimension.
Data frame : if output is a single column, returns a vector instead of a data frame.
$ is a shorthand operator, where x$y is equivalent to x[["y", exact = FALSE]] . It’s often used to access variables in a data frame, as in mtcars$cyl or diamonds$carat .
One common mistake with $ is to try and use it when you have the name of a column stored in a variable:
There’s one important difference between $ and [[ . $ does partial matching:
If you want to avoid this behaviour you can set the global option warnPartialMatchDollar to TRUE . Use with caution: it may affect behaviour in other code you have loaded (e.g., from a package).
Missing/out of bounds indices
[ and [[ differ slightly in their behaviour when the index is out of bounds (OOB), for example, when you try to extract the fifth element of a length four vector, or subset a vector with NA or NULL :
The following table summarises the results of subsetting atomic vectors and lists with [ and [[ and different types of OOB value.
If the input vector is named, then the names of OOB, missing, or NULL components will be "<NA>" .
- Given a linear model, e.g., mod <- lm(mpg ~ wt, data = mtcars) , extract the residual degrees of freedom. Extract the R squared from the model summary ( summary(mod) )
Subsetting and assignment
All subsetting operators can be combined with assignment to modify selected values of the input vector.
Subsetting with nothing can be useful in conjunction with assignment because it will preserve the original object class and structure. Compare the following two expressions. In the first, mtcars will remain as a data frame. In the second, mtcars will become a list.
With lists, you can use subsetting + assignment + NULL to remove components from a list. To add a literal NULL to a list, use [ and list(NULL) :
Applications
The basic principles described above give rise to a wide variety of useful applications. Some of the most important are described below. Many of these basic techniques are wrapped up into more concise functions (e.g., subset() , merge() , plyr::arrange() ), but it is useful to understand how they are implemented with basic subsetting. This will allow you to adapt to new situations that are not dealt with by existing functions.
Lookup tables (character subsetting)
Character matching provides a powerful way to make lookup tables. Say you want to convert abbreviations:
If you don’t want names in the result, use unname() to remove them.
Matching and merging by hand (integer subsetting)
You may have a more complicated lookup table which has multiple columns of information. Suppose we have a vector of integer grades, and a table that describes their properties:
We want to duplicate the info table so that we have a row for each value in grades . We can do this in two ways, either using match() and integer subsetting, or rownames() and character subsetting:
If you have multiple columns to match on, you’ll need to first collapse them to a single column (with interaction() , paste() , or plyr::id() ). You can also use merge() or plyr::join() , which do the same thing for you — read the source code to see how.
Random samples/bootstrap (integer subsetting)
You can use integer indices to perform random sampling or bootstrapping of a vector or data frame. sample() generates a vector of indices, then subsetting to access the values:
The arguments of sample() control the number of samples to extract, and whether sampling is performed with or without replacement.
Ordering (integer subsetting)
order() takes a vector as input and returns an integer vector describing how the subsetted vector should be ordered:
To break ties, you can supply additional variables to order() , and you can change from ascending to descending order using decreasing = TRUE . By default, any missing values will be put at the end of the vector; however, you can remove them with na.last = NA or put at the front with na.last = FALSE .
For two or more dimensions, order() and integer subsetting makes it easy to order either the rows or columns of an object:
More concise, but less flexible, functions are available for sorting vectors, sort() , and data frames, plyr::arrange() .
Expanding aggregated counts (integer subsetting)
Sometimes you get a data frame where identical rows have been collapsed into one and a count column has been added. rep() and integer subsetting make it easy to uncollapse the data by subsetting with a repeated row index:
Removing columns from data frames (character subsetting)
There are two ways to remove columns from a data frame. You can set individual columns to NULL:
Or you can subset to return only the columns you want:
If you know the columns you don’t want, use set operations to work out which colums to keep:
Selecting rows based on a condition (logical subsetting)
Because it allows you to easily combine conditions from multiple columns, logical subsetting is probably the most commonly used technique for extracting rows out of a data frame.
Remember to use the vector boolean operators & and | , not the short-circuiting scalar operators && and || which are more useful inside if statements. Don’t forget De Morgan’s laws , which can be useful to simplify negations:
- !(X & Y) is the same as !X | !Y
- !(X | Y) is the same as !X & !Y
For example, !(X & !(Y | Z)) simplifies to !X | !!(Y|Z) , and then to !X | Y | Z .
subset() is a specialised shorthand function for subsetting data frames, and saves some typing because you don’t need to repeat the name of the data frame. You’ll learn how it works in non-standard evaluation .
Boolean algebra vs. sets (logical & integer subsetting)
It’s useful to be aware of the natural equivalence between set operations (integer subsetting) and boolean algebra (logical subsetting). Using set operations is more effective when:
You want to find the first (or last) TRUE .
You have very few TRUE s and very many FALSE s; a set representation may be faster and require less storage.
which() allows you to convert a boolean representation to an integer representation. There’s no reverse operation in base R but we can easily create one:
Let’s create two logical vectors and their integer equivalents and then explore the relationship between boolean and set operations.
When first learning subsetting, a common mistake is to use x[which(y)] instead of x[y] . Here the which() achieves nothing: it switches from logical to integer subsetting but the result will be exactly the same. In more general cases, there are two important differences. First, when the logical vector contains NA, logical subsetting replaces these values by NA while which() drops these values. Second, x[-which(y)] is not equivalent to x[!y] : if y is all FALSE, which(y) will be integer(0) and -integer(0) is still integer(0) , so you’ll get no values, instead of all values. In general, avoid switching from logical to integer subsetting unless you want, for example, the first or last TRUE value.
How would you randomly permute the columns of a data frame? (This is an important technique in random forests.) Can you simultaneously permute the rows and columns in one step?
How would you select a random sample of m rows from a data frame? What if the sample had to be contiguous (i.e., with an initial row, a final row, and every row in between)?
How could you put the columns in a data frame in alphabetical order?
Positive integers select elements at specific positions, negative integers drop elements; logical vectors keep elements at positions corresponding to TRUE ; character vectors select elements with matching names.
[ selects sub-lists. It always returns a list; if you use it with a single positive integer, it returns a list of length one. [[ selects an element within a list. $ is a convenient shorthand: x$y is equivalent to x[["y"]] .
Use drop = FALSE if you are subsetting a matrix, array, or data frame and you want to preserve the original dimensions. You should almost always use it when subsetting inside a function.
If x is a matrix, x[] <- 0 will replace every element with 0, keeping the same number of rows and columns. x <- 0 completely replaces the matrix with the value 0.
A named character vector can act as a simple lookup table: c(x = 1, y = 2, z = 3)[c("y", "z", "x")]
© Hadley Wickham. Powered by jekyll , knitr , and pandoc . Source available on github .
More R Subscript
This page has the following sections:
Vector Matrix and Data Frame List Replacement Matrix and Array Resources
Figures 1 and 2 show the results of six cases of subscripting a particular vector that can be defined with:
This particular vector is numeric, but subscripting is the same no matter what type of vector it is.
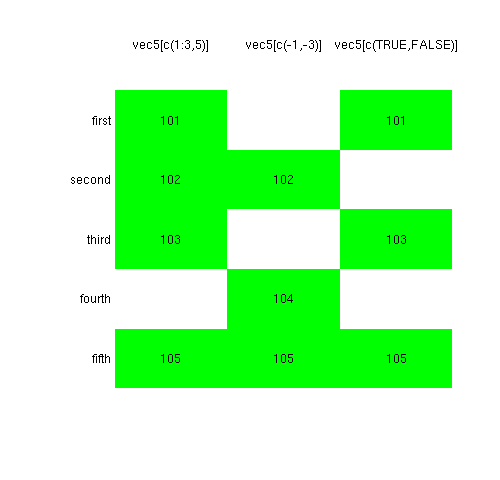
Positive numbers
When you subscript with positive numbers, you are saying which elements you want to keep.
There are two ways in which Figure 1 is misleading for this case:
- the order need not be the same as the original.
For example
is different from
- subscripts can be repeated.
Negative numbers
When you subscript with negative numbers, you are saying which elements you don’t want to keep. The order of elements in the resulting vector is always the same as in the original — there are just some elements gone.
Logical values
When you subscript with a logical vector, you are selecting the elements that correspond to TRUE . Unlike in Figure 1, this is often something like:
That is, the logical vector doing the subscripting is the same length as the original vector, and it is the result of some comparison operation.
A logical subscript is similar to a negative number subscript. They both leave the elements of the result in the same order as the original with some of the elements not there.
The example in Figure 1 is actually rather weird. Since the subscripting vector is shorter than the original vector, it is replicated to have the same length as the original. Such recycling is either handy or dangerous, depending on the context.
Character values
Subscripting with a character vector is just the same as subscripting with positive numbers. The difference is that you are specifying the elements to select using the names of the vector rather than the order within the vector.
This correspondence means that the order of the result need not be the same as in the original, and there can be repeated elements.
If there is no argument inside the square brackets, then all elements of the original vector are selected. This seems silly. But when we get to matrices and data frames, we’ll see that it is actually very useful.
Zero length
While it might seem like putting NULL inside the square brackets would be the same as putting nothing there, the results couldn’t be more different.
Subscripting with any zero length object produces a zero length result.
Matrix and Data Frame
A matrix and a data frame are subscripted exactly the same. A vector is conceptually a one-dimensional object while matrices and data frames are two-dimensional. So you need two subscripts for matrices and data frames. The rows and the columns are subscripted independently of each other, with a comma separating the subscripts. Here are some examples, where twodim can be either a matrix or a data frame:
When characters are used as subscripts, this will be referring to the row names or the column names.
If you subscript a matrix so that it has one row or one column, then the result is not a matrix — it is a plain vector. You can ensure that the result is a matrix by adding a drop=FALSE to the subscripting:
If you subscript only one column of a data frame, then the default behavior is to not get a data frame. That is almost always what you want. If not, then you can use the drop argument to subscripting as with matrices. If you subscript a single row of a data frame, you get a data frame.
You subscript lists just like vectors. Lists are vectors in the sense that counts for subscripting. Subscripting is the same no matter what type of vector it is.
There is another type of subscripting with lists — using double square brackets — that selects one component of the list.
selects the first component of the list. That is different from
which returns a list with one component.
The difference is a little bit subtle. But only a little bit.

If a list is a train of boxcars, then train[1] is the first boxcar and train[[1]] is what is inside the first boxcar. (I’m so jealous that I didn’t think of this analogy.)
The double bracket is going down inside and pulling out a piece. That piece might be a list, but it doesn’t have to be.
Let’s create a list and experiment.
A way to remember it is that subscripting with a single bracket can have length greater than one, so the result has to be a list. The result of the double bracket subscripting has whatever mode that particular component of the list has.
The example above gave a hint of another form of subscripting — the use of the $ operator.
This takes the name of the list on the left and the name of the component you want on the right. The result is the component.
The $ operator and the double square bracket are very similar. All of the following are doing the same thing:
A difference between the $ operator and double square brackets is that double square brackets can be used on all types of vectors, but $ is specific to lists.

Replacement
The discussion above only talks about extraction. However, if the subscripting is on the left of an assignment, then the values are replaced. That is, the original object has some of its elements modified.
changes vec5 so that 103 is its maximum value.
Matrix and Array
Matrices are actually a special case of arrays — a matrix is a two dimensional array. An array can have any dimension (that fits on the machine).
When you subscript an array, you need to have as many subscripts as there are dimensions in the array. So you might subscript a three-dimensional array like:
The “need” just above is a lie. You can also give just one subscript. There are two ways of giving just one subscript:
Vector subscript
An array (including a matrix) is really just one long vector that has a dim attribute that says how to wrap its values into a higher dimensional shape.
You are free to subscript as if the object were a plain vector.
There is a danger that you can do this accidentally. For example, you might say
when you mean to say
The difference in the command is tiny, the difference in the result can be dramatic.
You can also subscript a data frame with a single vector, but the result is much different than with a matrix. A data frame is really a list with a component for each column. So subscripting a data frame with a single vector means you are selecting columns.
Matrix subscript (advanced)
You can subscript an n -dimensional array with a matrix that has n columns. Each row contains the indices in each dimension for the location to be subscripted. The result is a vector with length equal to the number of rows in the subscripting matrix.
In modern versions of R the subscripting matrix can be either positive numbers or character. However, in older versions only numeric subscripting is supported.
Suppose that we have a matrix called Mat and we want the value from a random column for each row. (Why would we want to do that? I don’t know either.) The following command does that:
Circle 8 of The R Inferno contains some ways to be fooled when subscripting.
Back to top level of Impatient R
The train picture is by Mathias25 via stock.xchng

- About Burns Statistics
- Tao Te Programming
- The R Inferno
- Presentations
- Working Papers
- Contact Details
- Favorite pages
- Probably the most useful R function I’ve ever written
- Source for the marketAgent R package
- Review of ‘Advanced R’ by Hadley Wickham
- The wings of a programmer
- An experience of EARL
- Bad Science
- Data Science London
- Decision Science News
- FlowingData
- More or Less
- Portfolio Probe
- Science Corgi
- Significance Magazine
- Simply Statistics
- Statistical Modeling, Causal Inference, and Social Science
- Stats with Cats
- The numbers guy
- The Statistics Forum
- Understanding Uncertainty
Latest Tweets
- Book review
- Programming
- Uncategorized
- August 2016
- January 2015
- August 2014
- February 2014
- January 2014
- December 2013
- November 2013
- October 2013
- February 2013
- January 2013

R Error missing values not allowed in subscripted assignments (2 Examples)
In this R tutorial you’ll learn how to deal with the “Error in X : missing values are not allowed in subscripted assignments of data frames”.
Creation of Example Data
my_df <- data.frame(var1 = c(NA, "a", NA, "a", "b"), # Create example data var2 = 11:15) my_df # Print example data # var1 var2 # 1 <NA> 11 # 2 a 12 # 3 <NA> 13 # 4 a 14 # 5 b 15
Example 1: Reproduce the Error – missing values are not allowed in subscripted assignments of data frames
my_df[my_df$var1 == "a", ]$var2 <- 100 # Try to replace values # Error in `[<-.data.frame`(`*tmp*`, my_df$var1 == "a", , value = list(var1 = c(NA, : # missing values are not allowed in subscripted assignments of data frames
Example 2: Fix the Error – missing values are not allowed in subscripted assignments of data frames
my_df[!is.na(my_df$var1) & my_df$var1 == "a", ]$var2 <- 100 # Properly replace values my_df # Print updated data # var1 var2 # 1 <NA> 11 # 2 a 100 # 3 <NA> 13 # 4 a 100 # 5 b 15
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Post Comment
Draw User-Defined Axis Ticks Using axis() Function in R (Example Code)
Get number of missing values by group in r (example code), how to supress warning messages upon loading add-on packages in r (example code).
NAs are not allowed in subscripted assignments
I'm trying to run the following code , but have error. Please suggest!
df$ppiedu2[df$ppi_mm_educ==12]<- df$education[df$ppi_mm_educ==12]
Error in df$ppiedu2[df$ppi_mm_educ == 12] <- df$education[df$ppi_mm_educ == : NAs are not allowed in subscripted assignments
What is the result of
If that returns TRUE, then at least one of the subscript values you are passing is NA.
below is my dataset, once ppi_mm_educ==12, replace ppiedu2 with education like df$ppiedu2[df$ppi_mm_educ==12]<- df$education[df$ppi_mm_educ==12]
Is this what you are trying to do?
I got the right way without error, and thanks for your help!!
df$ppiedu2[df$ppi_mm_educ==12 & !is.na(df$ppi_mm_educ)]<- df$education[df$ppi_mm_educ==12 & !is.na(df$ppi_mm_educ)]
This topic was automatically closed 7 days after the last reply. New replies are no longer allowed. If you have a query related to it or one of the replies, start a new topic and refer back with a link.
Search code, repositories, users, issues, pull requests...
Provide feedback.
We read every piece of feedback, and take your input very seriously.
Saved searches
Use saved searches to filter your results more quickly.
To see all available qualifiers, see our documentation .
- Notifications
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement . We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Computing nearest neighbor graph: Error in m[match(oldnodes,m)] <-1:(N-1): NAs are not allowed in sybscripted assignments. #510
krigia commented Feb 20, 2023
GeorgescuC commented Feb 22, 2023
Sorry, something went wrong.
krigia commented Feb 28, 2023
GeorgescuC commented Mar 2, 2023
Krigia commented mar 2, 2023, krigia commented mar 24, 2023 • edited, georgescuc commented mar 24, 2023, krigia commented mar 24, 2023, georgescuc commented apr 7, 2023, georgescuc commented apr 24, 2023.
No branches or pull requests

Howdy, Stranger!
It looks like you're new here. If you want to get involved, click one of these buttons!
Quick Links
- Recent Discussions
- 8.4K All Categories
- 5K OpenSesame
- 1.8K JASP & BayesFactor
- 100 Expyriment
- 101 Mousetrap
- 25 DataMatrix
- 149 Miscellaneous

NAs are not allowed in subscripted assignments
Hi! I am analyzing my data from a web experiment, and when I try to run this command:
I get the following error message:
as you can see, I have eliminated trials with no movement. Any help would be greatly appreciated!
I think you have to remove all the NA values in your data set. Are you sure you have removed all of the NA variables? If so, there should be no problem. 😊
Despite removing all positions as spesified above like a.g.leontiev did I'm getting the same error. Can you explain how remove all the NAs from the mousetrap object @GoSugar please? Any help would be greatly appreciated!
I also would speculate that the problem is due to missing values (NAs), probably missing values that are already occurring in the raw dataset (df_sst).
Usually, mt_import_mousetrap should to a good job at checking for missing values and throwing warning messages if there are any. Do you get any warning messages after running mt_import_mousetrap with your dataset and, if so, could you post them here?
I also have an (unrelated) question. Is there a way to calculate measures not over ALL trials, but let's say first half of the trials? Thank you so much!
Great to hear things are running smoothly now!
Currently, there is no option in mousetrap that can calculate measures for only parts of the trial. However, depending on what measure you are interested in, functions like mt_deviations and mt_derivatives calculate deviatons from the idealized trajectory and velocity/accelerations for each time point, so you might use this as a starting point if you want to calculate a measure for a certain time point / part of the trial.
Hi Pascal thanks for getting back to me. This task is a flanker task run in Qualtrics, where the participants have to press start at the bottom of the screen then click on the top left or right depending which way the middle arrow is facing. I had to do a lot of preprocessing to get it into a format that is importable to mousetrap. I'm having issues with my code. When I just do mt_align there are no errors but the agregate trajectories aren't right:
mt_data <- mt_import_wide(mt_data1)
mt_data <- mt_count(mt_data, save_as = "data")
# Look at number of of recorded positions
table(mt_data$data$nobs)
# Only keep trials with more than 2 recorded positions
mt_data <- mt_subset(mt_data, nobs > 2)
# Calculate variance of positions for each trial
mt_data$data$pos_var <- apply(mt_data$trajectories[,,"xpos"],1,var,na.rm=TRUE) + apply(mt_data$trajectories[,,"ypos"],1,var,na.rm=TRUE)
# Check if there are trials with 0 variance (i.e., all positions are identical)
table(mt_data$data$pos_var==0)
# Only keep trials where positions varied
mt_data <- mt_subset(mt_data, pos_var>0)
mt_data <- mt_derivatives(mt_data)
mt_data<-mt_measures(mt_data, use = "trajectories",save_as="measures",
dimensions = c("xpos", "ypos"), timestamps = "timestamps",
verbose = FALSE)
mt_data <- mt_time_normalize(mt_data)
mt_data <- mt_spatialize(mt_data)
mt_data$trajectories[,,"ypos"] <- (-mt_data$trajectories[,,"ypos"])
mtdata1<-mt_align(mt_data)
mt_plot(mtdata1, use="trajectories", color="subj")
mt_plot_aggregate(mt_data, use="sp_trajectories", color="condition")
However as you can see this isn't right. If I just do mt_plot_aggregate(mt_data, use="trajectories", color="condition")
If I try to specify the start and end to something meaningful from the first figure :
mtdata1<-mt_align_start_end(mt_data, use = "trajectories", save_as = use,
dimensions = c("xpos", "ypos"), start = c(1250, -1250), end = c(0,0),
verbose = FALSE)
I get the following error:
Error in create_results(data = data, results = trajectories, use = use, : object 'use' not found
In addition: Warning message: In mt_align_start_end(mt_data, use = "trajectories", save_as = use, : NaN/Inf/-Inf values returned in some trials for the dimension xpos as their start and end coordinate was identical .
Where as if I just specify mt_align further mtdata<-mt_align( mt_data, use = "trajectories", save_as = use, dimensions = c("xpos", "ypos"), coordinates = c(-1200,1200,-2500,0), align_start = TRUE, align_end = TRUE, align_side = "left", verbose = FALSE)
Error in trajectories[flip, , dimensions[1]] <- ((trajectories[flip, , : NAs are not allowed in subscripted assignments
How can I still have NAs when I've removed trials without variance or <2positions? And why does mt_align work but not mt_align_start(end)
I really appreciate all your help. Thanks so much,
I meant not in the first part of a trial, but let's say first 100 of ALL trials :)
Oh Sorry there isn't an obvious error on import
>mt_data <- mt_import_wide(mt_data1)
No mt_id_label provided. A new trial identifying variable called mt_id was created.
No pos_ids provided. The following variables were found using grep:
397 variables found for timestamps.
397 variables found for xpos.
397 variables found for ypos.
Regarding @a.g.leontiev 's question:
I am not completely sure what you mean with "I meant not in the first part of a trial, but let's say first 100 of ALL trials".
Do you mean you would only like to select a subset of trials for your analysis? If so, you could do this either using the basic R subset function before importing your data into mousetrap or using mt_subset afterwards. Assuming you have a variable that is your trial count (e.g., "count_trial"), the command would simply look like:
raw_data <- subset(raw_data, count_trial<=100)
mt_data a- mt_subset(mt_data, count_trial<=100)
Regarding @P_Barr 's question:
I think that there are still some problematic trials in your dataset that need to be excluded. Specifically, there is the above warning that you quote:
The warning suggest that in some trials the start and end point is identical which, if I understand your setup correctly, should not be possible (if participants click a start button at the bottom and then one of the response buttons at the top of the screen). One explanation for this that I can think of is that your task uses a time limit after which a trial is automatically aborted (i.e., there are trials without response possible). If so, these trials need to be excluded before importing them into mousetrap.
You are correct its not possible to have the same start and end position as they have to click start in the bottom center of the screen and then a left or right button at the top corners.
I don't think we included a timeout- participants are forced to click a response button to see the next trial. We did include an error message after they clicked a response button if they started too late (>800ms) or responded too late (>5000ms). I have excluded these prior to import (wide) and now the trajectories seem to automatically have an aligned start and end but I'm still having weird aggregate plotting issues:
mt_data1<-read.csv ("MT_data5.csv", header = TRUE, sep = ',')
# remove too long/timeout responses prior to import( https://forum.cogsci.nl/discussion/comment/20455#Comment_20455)
mt_data1 <- mt_data1[ which(mt_data1$init.time <800 & mt_data1$response_latency2<5000), ]
#import to MT
mt_data<-mt_measures(mt_data, use = "trajectories",save_as="measures", dimensions = c("xpos", "ypos"), timestamps = "timestamps", verbose = FALSE)
mt_plot(mt_data, use="trajectories", color="condition")
mt_plot_aggregate(mt_data, use="sp_trajectories",color="condition")
mt_plot_aggregate(mt_data, use="trajectories",+ color="condition")
Warning message:
In mt_reshape(data = data, use = use, use_variables = use_variables, :
Trajectories differ in the number of logs. Aggregate trajectory data may be incorrect
If I try and do align_start_end (which I don't think I need to as it seems to be aligned) I get the same error message:
mtdata1<-mt_align_start_end(mt_data, use = "trajectories", save_as = use,dimensions = c("xpos", "ypos"), start = c(1250, -1250), end = c(0,0), verbose = FALSE)
Error in create_results(data = data, results = trajectories, use = use, : object 'use' not found
In addition: Warning message:
As always I really appreciate your help and any ideas
I have the following ideas regarding the issues you mentioned above:
1) Flipping trajectories
In your preprocessing code, I did not see a step where you flip trajectories that ended on the right option to the left. If you do not do this, the resulting aggregate trajectory plot might be misleading as it aggregates trajectories that end on the left and end on the right option.
2) Aggregate trajectory plot:
To create the aggregate trajectory plot, you use:
mt_plot_aggregate(mt_data, use="trajectories", color="condition")
However, when plotting aggregate trajectories, you usually cannot use the raw trajectories as the number of recorded positions varies between trajectories (which is also why you get the warning message you quoted above: "Trajectories differ in the number of logs. Aggregate trajectory data may be incorrect"). Therefore, the resulting plot looks quite weird. Instead, you could use the time-normalized trajectories:
mt_plot_aggregate(mt_data, use="tn_trajectories", color="condition")
3) Start end alignment
The warning message you quoted "NaN/Inf/-Inf values returned in some trials for the dimension xpos as their start and end coordinate was identical." suggests that in some trials the x coordinate of the start and end position was identical (the y coordinate might vary). In this case, the function cannot normalize the trajectory start and end points. I don't know if an identical start and end x coordinate is possible in your setup, but if so, it might not make sense to use this function.
I am unfortunately having this issue again:#### 100 ms ####
Any help would be greatly appreciated @Pascal . As you can see above, I tried removing trials with no movement, but I still get the same problem.
I have a questions to better understand the issue. In your code and output it says after running mt_import_mousetrap:
Have you inspected these warning messages and could post them here?
The error messages further down in your code (" NAs are not allowed in subscripted assignments") suggest that some trajectories might have a different number of recorded x/y-positions. This might already be in issue in the raw data, which could be raised by the wawrning messages in mt_import_mousetrap.
@Pascal Thank you for the prompt response. Here is the output, and I think you are correct, but how we get rid of it later so that we can still calculate stopping distance for a given time delay?
if you only want to exclude the probelamtic trials, one "quick and dirty" solution could be to just exclude the ids that were reported directly after mt_import_mousetrap with mt_subset:
However, it would probably be better to explore why this warning occurs by closer inspecting the raw data and the tool that was used to collect that mouse-tracking data.
Hi @Pascal ,
I hope you are well and safe. :)
Then I get a warning message:
With the above, I get the following warning:
then I load the behavioural data:
I then add columns to the dataset:
When I try to plot the trajectories, there is a third condition (i.e. NA) which should not be there:
When I try to plot according to TypeReader, it shows NA instead of Above.:
in the plot above, NA is no longer present, but neither is Above
My apologies for such lengthy post!
This is my first time doing mousetracking analysis and I am not super skilled at R.
Hi @Pascal ,
This is my first time analysing mousetracking data, and I am having issues with NA values in my dataset. Below is my step-by-step approach to the analysis as I don't know what I am doing wrong.
This is my first time doing mousetracking analysis, and I am not super skilled at R.
before answering your question, I would like to point out that I am not familiar with the Gorilla data format so I can probably only give you more general advice on how to format your data in a way that they are processed by mouestrap correctly.
The general logic when you import data into mousetrap is that you import all the data you need already in the mt_import_long step and do not add variables to the data part afterwards (I am not saying that this is not possible but if you do it the other way this should ensure all data are assigned to the correct trajectory).
In your case this would mean that you should merge gorilla.mt and gorilla.response.attempts datasets (using the trial identifying variable(s)). In the resulting dataset, each response variable would then be stretched across the complete trial (with a constant value per trial). This dataset could then be imported with mt_import_long in the same way you are doing it above.
Regarding the aggregate plots, I can only speculate but I would guess that you have both trajectories in the data that end on a option on the left and that end on an option on the right which is why the aggregate looks weird and somewhere in between. Usually you you remap all trajectories before plotting so that all trajectories end on the same response option side.
Hope this helps!
Thank you for your response!
I have followed your advice and added the additional columns prior to importing the data as a mousetrap object. However, I am having some odd issues now.
Below is the structure of the mousetracking and behavioural data together:
There are 36 trials per participant, so not sure if the below make sense:
this is the plot i get:
Why does it work the second time around, but not the first?
oops, I accidentally didn't finish the above.
I have already cleaned the global environment and restarted R several times, but the above keeps happening.
Thank you once again, @Pascal :) I really appreciate your help!
Have a nice week!
regarding the question if nrow(gorilla.mt$data) = 1692 makes sense?
If you have 36 trials per participants, then you should have 47 participants in there as nrow should be the total number of trials in the data (i.e., number of participants * number of trials / participant).
Regarding the NAs: this is in general no reason to worry as mousetrap internally represents the trajectories in an array and has to fill up shorter trajectories with NAs up to the length of the longest trajectory in the data.
Regarding the odd plot: I think the explanation is the following: mt_remap_symmetric only works if you have a centered coordinate system - see documentation, e.g., here: http://pascalkieslich.github.io/mousetrap/reference/mt_remap_symmetric.html
This does not seem to be the case in your raw data. However, once you align your trajectories with mt_align_start(gorilla.mt), the function actually works (at least with regard to mirroring all trajectories that end on the right option to the left). So you should be able to reproduce the desired behavior when running the code once but first calling mt_align_start and then mt_remap_symmtetric.
Thank you so much for your swift reply.
It is working now! Thank you for clarifying! It all makes sense now!

Secure Your Spot in Our PCA Online Course Starting on April 02 (Click for More Info)
Handling Errors & Warnings in R | List of Typical Messages & How to Solve
This page explains some of the most common error and warning messages in the R programming language .
Below, you can find a list of typical errors and warnings. When clicking on the bullet points of the list, you are headed to detailed instructions on how to deal with the corresponding error or warning message.
You may use the list as cheat sheet whenever you are facing an error or warning message in R.
Let’s dive into the examples!

List of Typical Errors & Warnings in R (+ Examples)
- [ reached getOption(“max.print”) — omitted X entries ]
- `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
- `summarise()` has grouped output by ‘X’. You can override using the `.groups` argument.
- Don’t know how to automatically pick scale for object of type standardGeneric. Defaulting to continuous.
- Error: ‘\U’ used without hex digits in character string starting “”C:\U”
- Error: `data` must be a data frame, or other object coercible by `fortify()`, not an S3 object with class uneval
- Error: (list) object cannot be coerced to type ‘double’
- Error: ‘R’ is an unrecognized escape in character string starting “”C:R”
- Error: Aesthetics must be either length 1 or the same as the data
- Error: attempt to apply non-function
- Error: Can’t rename columns that don’t exist.
- Error: cannot allocate vector of size X Gb
- Error: Continuous value supplied to discrete scale
- Error: Discrete value supplied to continuous scale
- Error: geom_point requires the following missing aesthetics: x, y
- Error: Insufficient values in manual scale. X needed but only Y provided.
- Error: Invalid input: date_trans works with objects of class Date only
- Error: JAVA_HOME cannot be determined from the Registry
- Error: Mapping should be created with `aes()` or `aes_()`.
- Error: object X not found
- Error: stat_count() must not be used with a y aesthetic.
- Error: StatBin requires a continuous x variable: the x variable is discrete.Perhaps you want stat=”count”?
- Error: unexpected ‘,’ in “,”
- Error: unexpected ‘}’ in X
- Error: unexpected ‘=’ in “=”
- Error: unexpected ‘)’ in “)”
- Error: unexpected ‘else’ in “else”
- Error: unexpected input in X
- Error: unexpected numeric constant in X
- Error: unexpected SPECIAL in X
- Error: unexpected string constant in X
- Error: unexpected symbol in X
- Error: X must only be used inside dplyr verbs.
- Error in .Call.graphics : invalid graphics state
- Error in .subset(x, j) : invalid subscript type ‘list’
- Error in `levels<-`(`*tmp*`, value = as.character(levels)) : factor level is duplicated
- Error in aggregate.data.frame(as.data.frame(x), …) : arguments must have same length
- Error in apply(data) : dim(X) must have a positive length
- Error in as.Date.numeric(X) : ‘origin’ must be supplied
- Error in as.POSIXlt.character(x, tz, …) : character string is not in a standard unambiguous format
- Error in barplot.default(X) : ‘height’ must be a vector or a matrix
- Error in charToDate(x) : character string is not in a standard unambiguous format
- Error in colMeans(x, na.rm = TRUE) : ‘x’ must be numeric
- Error in contrasts : contrasts can be applied only to factors with 2 or more levels
- Error in cut.default : ‘breaks’ are not unique
- Error in do_one(nmeth) : NA/NaN/Inf in foreign function call (arg 1)
- Error in eval(predvars, data, env) : numeric ‘envir’ arg not of length one
- Error in file(file, “rt”) : cannot open the connection
- Error in file(file, “rt”) : invalid ‘description’ argument
- Error in fix.by(by.y, y) : ‘by’ must specify a uniquely valid column
- Error in FUN(X[[i]], …) : object ‘X’ not found
- Error in FUN : invalid ‘type’ (character) of argument
- Error in FUN : invalid ‘type’ (list) of argument
- Error in hist.default(X) : ‘x’ must be numeric
- Error in if (NA) { : missing value where TRUE/FALSE needed
- Error in library(“X”) : there is no package called ‘X’
- Error in lm.fit(x, y, offset = offset, singular.ok = singular.ok, …) : 0 (non-NA) cases
- Error in lm.fit(x, y, offset = offset, singular.ok = singular.ok, …) : NA/NaN/Inf in ‘x’
- Error in load(“X.rds”) : bad restore file magic number (file may be corrupted) — no data loaded
- Error in max.item – min.item : non-numeric argument to binary operator
- Error in model.frame.default(Terms, newdata, na.action = na.action, xlev = object$xlevels) : ‘data’ must be a data.frame, environment, or list
- Error in model.frame.default(Terms, newdata, na.action = na.action, xlev = object$xlevels) : factor X has new levels Y
- Error in names(X) : ‘names’ attribute must be the same length as the vector
- Error in Ops.data.frame() : only defined for equally-sized data frames
- Error in parse(text) : <text>:1:2: unexpected symbol
- Error in plot.new() : figure margins too large
- Error in plot.window(…) : need finite ‘xlim’ values
- Error in plot.xy(xy.coords(x, y), type = type, …) : plot.new has not been called yet
- Error in rbind(deparse.level, …) : numbers of columns of arguments do not match
- Error in read.table : more columns than column names
- Error in read.table(file = file, header = header, sep = sep, quote = quote, : duplicate ‘row.names’ are not allowed
- Error in rep(X) : invalid ‘times’ argument
- Error in rowSums & colSums – ‘x’ must be an array of at least two dimensions
- Error in scan : line 1 did not have X elements
- Error in setwd(X) : cannot change working directory
- Error in solve.default(X) : Lapack routine dgesv: system is exactly singular
- Error in sort.int(x, na.last = na.last, decreasing = decreasing, …) : ‘x’ must be atomic
- Error in stripchart.default(x1, …) : invalid plotting method
- Error in strsplit(X) : non-character argument
- Error in unique.default(x, nmax = nmax) : unique() applies only to vectors
- Error in UseMethod(“predict”) : no applicable method for ‘predict’ applied to an object of class “c(‘double’, ‘numeric’)”
- Error in X : $ operator is invalid for atomic vectors
- Error in X : argument is of length zero
- Error in X : arguments imply differing number of rows
- Error in X : attempt to select less than one element in get1index real
- Error in X : could not find function X
- Error in X : missing values are not allowed in subscripted assignments of data frames
- Error in X : incorrect number of dimensions
- Error in X : incorrect number of subscripts on matrix
- Error in X : invalid (do_set) left-hand side to assignment
- Error in X : invalid (NULL) left side of assignment
- Error in X : non-numeric argument to binary operator
- Error in X : object not interpretable as a factor
- Error in X : object of type ‘closure’ is not subsettable
- Error in X : replacement has Y rows, data has Z
- Error in X : requires numeric/complex matrix/vector arguments
- Error in X : subscript out of bounds
- Error in X : target of assignment expands to non-language object
- Error in X : undefined columns selected
- Error in X : unused argument
- Error in X %*% Y : non-conformable arguments
- Error in xy.coords(x, y, xlabel, ylabel, log) : ‘x’ and ‘y’ lengths differ
- geom_path: Each group consists of only one observation. Do you need to adjust the group aesthetic?
- Please select a CRAN mirror for use in this session
- Scale for ‘fill’ is already present. Adding another scale for ‘fill’, which will replace the existing scale.
- The following objects are masked from ‘package:X’
- The following objects are masked from X
- Use of `data$X` is discouraged. Use `X` instead
- Warning: cannot remove prior installation of package ‘X’
- Warning message: ‘newdata’ had X rows but variables found have Y rows
- Warning message: In cor(X) : the standard deviation is zero
- Warning message: In is.na(data) : is.na() applied to non-(list or vector) of type ‘closure’
- Warning message: In mean.default(X) : argument is not numeric or logical: returning NA
- Warning message: In min/max(X) : no non-missing arguments to min/max; returning Inf
- Warning message: In scan(file = file, what = what, sep = sep, quote = quote, dec = dec, : embedded nul(s) found in input
- Warning message: invalid factor level, NA generated
- Warning message: longer object length is not a multiple of shorter object length
- Warning message: NAs introduced by coercion
- Warning message: Removed X rows containing missing values
- Warning message: the condition has length > 1 and only the first element will be used
- Warning message in as.POSIXct.POSIXlt(x) : unknown timezone
- Warning message in Ops.factor : not meaningful for factors
- Warning message in read.table: incomplete final line found by readTableHeader
- Warning messages: In plot.window(…) : nonfinite axis limits [GScale(-inf,1,1, .); log=1]
- Warning messages: Removed 17 rows containing non-finite values (stat_bin).
Debugging in R – Some General Advice
Debugging in R can be a painful process. However, there are some useful tools and functions available that can be used to make the debugging more efficient.
One of these tools is the interactive debug mode , which is built into the RStudio IDE. This tool helps to find bugs by locating where the code is not working in the way you would expect, and can therefore be very helpful in case you are dealing with a more complex R code. You can read more about RStudio’s debug mode here .
Another useful method to handle errors and warnings is provided by the tryCatch function . This function can be used to check if an R code leads to an error or warning message, and it can be used to skip an error message in order to continue running the code. You can learn more about the application of the tryCatch function here .
In case you want to learn more about typical R programming error messages and the handling of these errors, you may also have a look at the following YouTube video. In the video, the speaker Andres Karjus explains some common error messages that beginners often get and how to quickly figure them out.
Note that there are usually different reasons why an error or warning message can occur. Please let me know in the comments in case the provided tips and tutorials on this page didn’t solve your problem.
Furthermore, please let me know in the comments in case the error or warning message you have problems with is not included in the previous list.
I’m looking forward to hearing from you in the comments!
Subscribe to the Statistics Globe Newsletter
Get regular updates on the latest tutorials, offers & news at Statistics Globe. I hate spam & you may opt out anytime: Privacy Policy .
32 Comments . Leave new
Error: grouping factors must have > 1 sampled level
Hey George,
Please have a look at this thread on Cross Validated. It seems to explain the error message.
Regards, Joachim
Error in make.names(col.names, unique = TRUE) : invalid multibyte string at ‘
Could you please share your code?
Hello Joachim,
May i know what does it mean getting this warning “There were 50 or more warnings (use warnings() to see the first 50)”? I am running a glmm analysis with the code something like this: model <-glmer (richness ~ var1 + var2 + var3+(1|habitat), family = "poisson", data=xxx) summary(model)
thanks in advance
Hey Esther,
This means that there have been many warnings after executing your code (i.e. more than 50). You may execute the following code to see them one-by-one:
Thanks Joachim,
Now if I run my glmm models I couldn’t get the results and got another warning messages which saying like this:
Warning messages: 1: In vcov.merMod(object, use.hessian = use.hessian) : variance-covariance matrix computed from finite-difference Hessian is not positive definite or contains NA values: falling back to var-cov estimated from RX 2: In vcov.merMod(object, correlation = correlation, sigm = sig) : variance-covariance matrix computed from finite-difference Hessian is not positive definite or contains NA values: falling back to var-cov estimated from RX >
When I got that warning messages, I’ve tried running my model using glmer.nb function instead glmer. May I know what the difference of these two functions? Can I actually do that, I mean change the function to glmer.nb to see the influence of the variables for my study?
Unfortunately, I’m not an expert on this topic and I have never experienced this problem myself. However, I found this thread on Stack Overflow , which seems to discuss this warning message. Maybe this helps?
Dear Joachim
I receive an error while I’m converting .CEL file to TXT and normalizing them :
after using this command : { raw.data <-ReadAffy(verbose = FALSE,filenames= f[i],cdfname="hgu133acdf") data.rma.norm = rma(raw.data)
it shows : Error in exprs(object)[index, , drop = FALSE] : subscript out of bounds ,I don't know what is the cause of this error: it's bcz my data is too long or maybe a bug is in my data or some thing else, It will be your kindness if guide me how I can fix this error ?
I do not have any experience with CEL files, but this error message is explained here .
I hope this helps!
Hi Joachim,
Have you ever seen anything like this?
I was plotting a simple gray bar graph and wanted to add some color. I originally ran this: ggplot(data=heartdata) + geom_bar(aes(x = fbs, fill = chol))
and it worked fine. I then played around trying to add some color and got this to work: ggplot(data=chol_sev) + geom_bar(aes(x = fbs, fill =..chol..))
I then closed R studio but when I reopened the script the color plot would no longer work. I now receive the below error. any ideas?
Error in `f()`: ! Aesthetics must be valid computed stats. Problematic aesthetic(s): fill = ..chol… Did you map your stat in the wrong layer? Run `rlang::last_error()` to see where the error occurred.
Please excuse the delayed response. I was on a long holiday, so unfortunately I wasn’t able to reply sooner. Still need help with your code?
(Error in { : task 1 failed – “‘list’ object cannot be coerced to type ‘integer'”)
I get this error
I found this error message during my R code using spatials data, and I don’t know how to resolve that. I don’t know if you help me with some idea: ” There were 12 warnings (use warnings() to see them) ”
Hey Nadine,
I’m sorry for the delayed response. I was on a long vacation, so unfortunately I wasn’t able to get back to you earlier. Do you still need help with your syntax?
Yes please !
I also have another question and I don’t know if you have time to direct me to it. I am looking to do a classification using deep learning for the identification of individual trees in a forest plot. I work on images taken with a multispectral sensor. I do it on the point cloud because the results obtained using the raster did not convince me. Normally you have to go through segmentation, alignment and classification. I did the segmentation with Dalpont algorithm and silva algorithm. But I can’t find the function I need for the two other steps (alignment and classification). I wonder if you have any idea about this.
Regarding your warning message, you could execute the following code in the RStudio console to see the actual warnings:
Regarding your additional question, unfortunately I’m not an expert on this topic. However, I run a Facebook discussion group where people can ask questions about R programming and statistics. Could you post your question there? This way, others can contribute/read as well: https://www.facebook.com/groups/statisticsglobe
Hi I am executing function in R studio and I got this error (Error in prettyNum(.Internal(format(x, trim, digits, nsmall, width, 3L, : invalid value 0 for ‘digits’ argument) I don’t know how to fix please help me if possible
I am not very familiar with that error. But my quick search shows that it pops up when there is a problem with the convergence of the glm algorithm. I found this StackOverflow thread . I hope it helps you.
Regards, Cansu
Hi, I have an excel with many columns, how can I convert all of them into numeric variables in Rstudio? error: the non-numeric argument to mathematical function I get this error
Hello Adrian,
Yes, you get this error because your variables are not numeric. I think our tutorial: Convert Data Frame Column to Numeric in R would help you with this.
Error in `[<-.data.frame`(`*tmp*`, is.na(df$Ozone), "Ozone", value = c(41L, : replacement has 153 rows, data has 37
Please have a look at this tutorial , it explains your error message.
When trying to run an ordered random forest, I got this error message: Error in check_Y(Y, X) : The number of outcomes does not equal the number of features. Check your data.
Hello Lagida,
The error message you’ve encountered typically indicates a discrepancy between the number of outcomes (target variable) and the number of features (predictor variables) in your dataset. You should verify that your data is properly formatted and organized to resolve this issue.
Here are some steps you can take to troubleshoot this problem:
Check your data: Ensure that the input data is structured correctly, with each row representing an observation and each column representing a feature. Ensure your target variable (Y) has the correct number of outcomes and your feature matrix (X) has the correct number of features.
Verify data types: Ensure that the data types of your variables are correct. For example, ensure that categorical variables are encoded as factors and numerical variables are encoded as numeric data types.
Check input parameters: Make sure that you have specified the correct input parameters for the ordered random forest function. This includes setting the appropriate values for the number of trees, the minimum number of samples per leaf, and any other tuning parameters specific to the function you are using.
Hi Jaochim,
I am unable to execute this piece of script in MacBook – I installed R studio Version 2023.09.1+494 (2023.09.1+494)
install.packages(“data.table”) # Install & load data.table library(“data.table”)
I am getting this error – Please do the needful.
There is a binary version available but the source version is later: binary source needs_compilation data.table 1.14.2 1.14.8 TRUE
Do you want to install from sources the package which needs compilation? (Yes/no/cancel) library(“data.table”) Error in install.packages : Unrecognized response “library(“data.table”)”
Thanks in advance.
Hello Chandru,
When you see the prompt Do you want to install from sources the package which needs compilation? (Yes/no/cancel), you need to type one of the options (Yes, no, cancel) and then press Enter. Typing Yes will compile the package from source, which can take a longer time and requires you to have the appropriate compiling tools installed on your MacBook (like Xcode command-line tools). Typing no will install the binary version, which is quicker and does not require compilation.
Only after the installation process has completed should you use the library function to load the package.
Best, Cansu
Hello Cansu,
Thanks for your reply, I tried with commnd ‘Yes’ and ‘No’ and even ‘cancel’ it did not work. Now, I need to find the code. I’ll get back to you if I need any help from you.
Much appreciated.
Okay let me know when you find it!
1: model fit failed for Fold01: maxdepth=1, mfinal=150 Error in imp[m, ] <- k[sort(row.names(k)), ] : number of items to replace is not a multiple of replacement length
Hello Somayyeh,
The error message you’re encountering seems to be related to an issue in a machine learning or statistical model fitting process, specifically in a cross-validation or similar iterative procedure.
Here’s a breakdown of the error message:
“model fit failed for Fold01: maxdepth=1, mfinal=150”: This indicates that the model fitting process failed for the first fold in a cross-validation or similar iterative process. maxdepth=1 suggests that the maximum depth of the tree (if it’s a tree-based model like a decision tree or a gradient boosting machine) was set to 1. mfinal=150 might indicate the number of trees or iterations for an ensemble method like Gradient Boosting or Random Forest.
“Error in imp[m, ] https://www.facebook.com/groups/statisticsglobe
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Post Comment

- All SEO Content SEO Course SEO Services Search for:
No products in the cart.
- Terms and Conditions
- Privacy Policy
- Refund Policy
- +94769416628
Ryter's Blog
Why nas are not permitted in subscripted assignments: understanding the restriction.
In this article, we will delve into the reasons why NAS (Network Attached Storage) devices are not permitted in subscripted assignments. We will explore the technical limitations and potential security risks that this restriction aims to address, and provide a comprehensive understanding of the rationale behind this rule. As organizations increasingly rely on networked storage solutions, it is crucial to understand the implications of using NAS in subscripted assignments and the reasons why it is not allowed.
What is the reasoning behind the restriction on NAS in subscripted assignments?
One of the primary reasons why NAS devices are not permitted in subscripted assignments is due to the potential security vulnerabilities they introduce. When a NAS device is used in a subscripted assignment, it creates an additional layer of access control and authorization, which may not align with the organization’s security policies. This can lead to unauthorized access to sensitive data and compromise the overall security posture of the network.
Furthermore, NAS devices often have their own operating systems and software that require regular updates and patches to mitigate vulnerabilities. When integrated into a subscripted assignment, managing the security of the NAS device becomes an added burden for the organization, which may not have the resources or expertise to do so effectively. As a result, prohibiting NAS in subscripted assignments helps to minimize the complexity of managing security risks associated with these devices.
What are the technical limitations that make NAS unsuitable for subscripted assignments?
NAS devices are designed to provide centralized storage and file sharing capabilities over a network. While they are efficient for these purposes, their architecture may not be optimized for use in subscripted assignments, particularly in a cloud-based or virtualized environment. The inherent latency and overhead of accessing data from a NAS device over the network can significantly impact the performance of subscripted assignments.
Additionally, NAS devices may not support the advanced features and protocols required for seamless integration with subscripted assignments. For example, the lack of support for certain network protocols or file systems may result in compatibility issues that hinder the functionality of subscripted assignments. By restricting the use of NAS in subscripted assignments, organizations can ensure that the underlying infrastructure is capable of meeting the performance and compatibility requirements of these assignments.
How does the restriction on NAS in subscripted assignments impact storage management?
The restriction on NAS in subscripted assignments reinforces the importance of centralized storage management and data governance. Instead of allowing individual users or departments to deploy NAS devices for their specific storage needs, organizations can consolidate their storage resources within a controlled environment. This centralized approach enables better oversight, data protection, and compliance with regulatory requirements.
Moreover, by standardizing the storage infrastructure for subscripted assignments, organizations can streamline storage management, backup, and disaster recovery processes. They can leverage scalable and resilient storage solutions that are inherently compatible with subscripted assignments, enabling seamless data access and availability. Ultimately, the restriction on NAS in subscripted assignments promotes a more cohesive and efficient approach to storage management within the organization.
What alternatives are available for meeting storage requirements in subscripted assignments?
While NAS devices are not permitted in subscripted assignments, organizations can explore alternative storage solutions that are better suited for these use cases. Cloud-based storage services, such as object storage and block storage, offer scalable and highly available storage options that integrate seamlessly with subscripted assignments. These services provide the flexibility and performance required to support a wide range of subscripted applications and workloads.
Additionally, organizations can consider deploying software-defined storage solutions that virtualize and abstract storage resources across the infrastructure. This approach enables the dynamic allocation and management of storage capacity for subscripted assignments, while maintaining the required levels of performance, security, and compliance. By embracing these alternatives, organizations can fulfill the storage requirements of subscripted assignments without compromising on security or performance.
In conclusion, understanding the restriction on NAS in subscripted assignments is essential for maintaining the security, performance, and manageability of the underlying infrastructure. By recognizing the potential security vulnerabilities, technical limitations, and storage management implications associated with using NAS devices in subscripted assignments, organizations can make informed decisions about their storage strategies. By exploring alternative storage solutions that align with the requirements of subscripted assignments, organizations can ensure that their storage infrastructure supports the evolving needs of modern applications and workloads.
Q: Can I use a NAS device for personal storage in a subscripted environment?
A: No, the restriction on NAS in subscripted assignments applies to both organizational and personal use. Personal NAS devices are not permitted in subscripted environments due to the potential security risks and management complexities they introduce.
Q: How does the use of NAS impact the performance of subscripted assignments?
A: NAS devices introduce additional network latency and overhead when accessing data, which can impact the performance of subscripted assignments. This can result in decreased responsiveness and throughput, especially for latency-sensitive applications.
Q: What security risks are associated with using NAS in subscripted assignments?
A: NAS devices may create additional access control and authorization layers that can be exploited by malicious actors. Furthermore, the inherent software and firmware of NAS devices may contain vulnerabilities that pose security risks to subscripted assignments.
Q: Are there any exceptions to the restriction on NAS in subscripted assignments?
A: In some cases, organizations may establish specific guidelines and controls for the use of NAS devices in subscripted assignments, based on stringent security and management requirements. However, such exceptions are rare and subject to rigorous scrutiny and compliance.
Q: How can organizations ensure compliance with the restriction on NAS in subscripted assignments?
A: Organizations can implement robust policies, technical controls, and monitoring mechanisms to enforce and track compliance with the restriction on NAS. This may include network isolation, access controls, and periodic audits of subscripted assignments.
Leave a Reply Cancel reply
Your email address will not be published. Required fields are marked *
Save my name, email, and website in this browser for the next time I comment.
Username or email address *
Password *
Remember me Log in
Lost your password?
Click one of our contacts below to chat on WhatsApp

IMAGES
VIDEO
COMMENTS
NAs mean Not Available values. This is very interesting. It seems that you can't combine integers with NA while subsetting and assigning only if you have more than one value on RHS... I.e. x[c(NA, 1)] <- 100 and x[NA] <- 100 will both work, while x[c(NA, 1)] <- 100:101 or x[NA] <- 100:101 won't. This is a valuable question as it gets at that ...
Introduction to Statistics is our premier online video course that teaches you all of the topics covered in introductory statistics.Get started with our course today.
How to Solve R Error: aesthetics must be either length 1 or the same as the data How to Solve R Error: continuous value supplied to discrete scale Go to the online courses page on R to learn more about coding in R for data science and machine learning.
Because when you split it up R doesn't have to infer what row an index of NA goes with; you're simply telling it to re-assign all the values in a particular column. - joran Apr 30, 2014 at 19:37
Subscribe my Newsletter for new blog posts, tips & new photos. Let's stay updated!
Subsetting. R's subsetting operators are powerful and fast. Mastery of subsetting allows you to succinctly express complex operations in a way that few other languages can match. Subsetting is hard to learn because you need to master a number of interrelated concepts: The three subsetting operators. The six types of subsetting.
So you need two subscripts for matrices and data frames. The rows and the columns are subscripted independently of each other, with a comma separating the subscripts. Here are some examples, where twodim can be either a matrix or a data frame: > twodim[1:3, 4:5] # rows 1, 2, 3, and columns 4, 5. > twodim[1:3, "id"] # rows 1, 2, 3, and column ...
Creation of Example Data. The following data will be used as basement for this R tutorial: x2 = 1:6) As you can see based on Table 1, the exemplifying data is a data frame comprising six rows and two columns called "x1" and "x2". The variable x1 has the numeric class and the variable x2 has the integer class.
Besides the usual numeric subscripts, R allows the use of character or logical values for subscripting. Subscripting operations are very fast and ... condition by using an appropriately subscripted object on the left-hand side. 6.5 Subscripting Matrices and Arrays 77 of an assignment statement. If we wanted to change the numbers in numsthat ...
Save my name, email, and website in this browser for the next time I comment.
NAs not allowed in subscripted assignments #52. Closed lmorett opened this issue May 14, 2018 · 12 comments Closed NAs not allowed in subscripted assignments #52. lmorett opened this issue May 14, 2018 · 12 comments Labels. bug. Comments. Copy link lmorett commented May 14, 2018.
Hi @andynkili,. Now that you mention the number of cells, the issue in this case might be R changing the notation of indices to scientific annotation, so index 100,000 (cell index for a distance table) becomes 1e5 which is then not recognized.
Hello, thank you for repalying! The original dataset has 614 rows. After omitting NA's, it goes down to 480. Here's a dataset before omitting NA's:
NAs are not allowed in subscripted assignments My code works well in local but got "Error: Input columns in `x` must be unique" when deploy to shinyapps.io FJCC October 3, 2020, 5:58pm
Assigning within a lapply "NAs are not allowed in subscripted assignments" Thanks in advance. r; machine-learning; random-forest; Share. Improve this question. Follow edited May 23, 2017 at 10:28. Community Bot. 1 1 1 silver badge. asked Dec 11, 2014 at 5:52. plr plr.
If you restart R, none of them should be loaded so it should be easier. If however they already are loaded when you restart your R session, you might have an .RData file in either your working directory or your home directory that gets reloaded with R and brings these issues back every time. Simply deleting it then restarting R should solve that.
Hi Pascal thanks for getting back to me. This task is a flanker task run in Qualtrics, where the participants have to press start at the bottom of the screen then click on the top left or right depending which way the middle arrow is facing.
Defaulting to continuous. Error: '\U' used without hex digits in character string starting ""C:\U". Error: `data` must be a data frame, or other object coercible by `fortify ()`, not an S3 object with class uneval. Error: (list) object cannot be coerced to type 'double'.
As already is inidicated in the line of code, the second condition does not run. And I do not know why. I checked here (NAs are not allowed in subscripted assignments) on this website to make sure I did not select NAs. However, appearently I miss something in the second line and I do not see what.
When integrated into a subscripted assignment, managing the security of the NAS device becomes an added burden for the organization, which may not have the resources or expertise to do so effectively. As a result, prohibiting NAS in subscripted assignments helps to minimize the complexity of managing security risks associated with these devices.
When trying to replace values, "missing values are not allowed in subscripted assignments of data frames" 5 R: Trouble using SMOTE package "invalid 'labels'" 0 Error: unused arguments using SMOTE. 1 Missing values are not allowed in subscripted assignments of data frames in MCMCglmm package ...